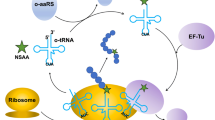
Abstract
Non-canonical amino acids as building blocks for protein biosynthesis facilitate modifications that go far beyond classical genetic engineering. Two complementary techniques allow either site-specific or residue-specific incorporation. Accordingly, the possible modifications range from atomic changes to the alteration of the overall biophysical characteristics of a protein. In this article, we present the state of the art of this new protein engineering approach.
Similar content being viewed by others
Literatur
Bogosian G, Violand BN, Dorward-King EJ et al. (1989) Biosynthesis and incorporation into protein of norleucine by Escherichia coli. J Biol Chem 254:531–539
Aoyagi Y, Sugahara T (1985) 2(S)-Aminohex-5-ynoic acid, an antimetabolite from Cortinarius claricolor var. Tenuipes. Phytochemistry 24:1835–1836
Ma SJ (2003) Unnatural amino acids in drug discovery. Chim Oggi 21:65–68
Bansal M, Ananthanarayanan VS (1988) The role of hydroxyproline in collagen folding: Conformational energy calculations on oligopeptides containing proline and hydroxyproline. Biopolymers 27:299–312
Johnson JA, Lu YY, van Deventer JA et al. (2010) Residuespecific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol 14:774–780
Furter R (1998) Expansion of the genetic code: Site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci 7:419–426
Young TS, Schultz PG (2010) Beyond the canonical 20 amino acids: Expanding the genetic lexicon. J Biol Chem 285:11039–11044
Mukai T, Yanagisawa T, Ohtake K et al. (2011) Geneticcode evolution for protein synthesis with non-natural amino acids. Biochem Biophys Res Commun 411:757–761
Johnson DBF, Xu J, Shen Z et al. (2011) RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol 7:779–786
Neumann H, Wang K, Davis L et al. (2010) Encoding multiple unnatural amino acids via evolution of a quadrupletdecoding ribosome. Nature 464:441–444
Chen H, Warfield L, Hahn S (2007) The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14:696–703
Service RF (2005) American Chemical Society Meeting: Unnatural amino acid could prove boon for protein therapeutics. Science 308:44
Mehl RA, Anderson JC, Santoro SW et al. (2003) Generation of a Bacterium with a 21 Amino Acid Genetic Code. J Am Chem Soc 125:935–939
Author information
Authors and Affiliations
Corresponding author
Additional information
Natascha Hotz, Tanja Marzluf und Birgit Wiltschi (v. l. n. r.)
Rights and permissions
About this article
Cite this article
Hotz, N., Marzluf, T. & Wiltschi, B. Protein Engineering mit nicht-kanonischen Aminosäuren. Biospektrum 18, 96–99 (2012). https://doi.org/10.1007/s12268-012-0146-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12268-012-0146-3