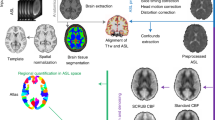
Abstract
Oxygenation-sensitive cardiovascular magnetic resonance (OS-CMR) is a novel, powerful tool for assessing coronary function in vivo. The data extraction and analysis however are labor-intensive. The objective of this study was to provide an automated approach for the extraction, visualization, and biomarker selection of OS-CMR images. We created a Python-based tool to automate extraction and export of raw patient data, featuring 3336 attributes per participant, into a template compatible with common data analytics frameworks, including the functionality to select predictive features for the given disease state. Each analysis was completed in about 2 min. The features selected by both ANOVA and MIC significantly outperformed (p < 0.001) the null set and complete set of features in two datasets, with mean AUROC scores of 0.89eatures f 0.94lete set of features in two datasets, with mean AUROC scores that our tool is suitable for automated data extraction and analysis of OS-CMR images.
Graphical Abstract






Similar content being viewed by others
Data Availability
All python code for the toolkit can be made available upon request. A sample of the data can also be provided on demand.
References
Khalique Z, Pennell DJ. What is CMR doing for patients today? Eur Heart J. 2018;39(4):266–70. https://doi.org/10.1093/eurheartj/ehx778.
Pennell DJ, Sechtem UP, Higgins CB, Manning WJ, Pohost GM, Rademakers FE,…Yucel EK. Clinical indications for cardiovascular magnetic resonance (CMR): consensus panel report. Eur Heart J, 2004;25(21), 1940–1965. https://doi.org/10.1016/j.ehj.2004.06.040
Hillier E, Friedrich MG. The potential of oxygenation-sensitive CMR in Heart failure. Curr Heart Fail Rep. 2021. https://doi.org/10.1007/s11897-021-00525-y.
Fischer K, Yamaji K, Luescher S, Ueki Y, Jung B, von Tengg-Kobligk H, Windecker S, Friedrich MG, Eberle B, Guensch DP. Feasibility of cardiovascular magnetic resonance to detect oxygenation deficits in patients with multi-vessel coronary artery disease triggered by breathing maneuvers. J Cardiovasc Magn Reson. 2018;20(1):31. https://doi.org/10.1186/s12968-018-0446-y.PMID:29730991;PMCID:PMC5937049.
Fischer K, Guensch DP, Jung B, King I, von Tengg-Kobligk H, Giannetti N, Eberle B, Friedrich MG. Insights into myocardial oxygenation and cardiovascular magnetic resonance tissue biomarkers in heart failure with preserved ejection fraction. Circ Heart Fail. 2022;15(4):e008903. https://doi.org/10.1161/CIRCHEARTFAILURE.121.008903.
Friedrich MG, Karamitsos TD. Oxygenation-sensitive cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2013;15(1):43. https://doi.org/10.1186/1532-429X-15-43.PMID:23706167;PMCID:PMC3681671.
Dharmakumar R, Qi X, Hong J, Wright GA. Detecting microcirculatory changes in blood oxygen state with steady-state free precession imaging. Magn Reson Med. 2006;55(6):1372–80. https://doi.org/10.1002/mrm.20911.
Hillier E, Covone J, Friedrich MG. Oxygenation-sensitive cardiac MRI with vasoactive breathing maneuvers for the non-invasive assessment of coronary microvascular dysfunction. JoVE (Journal of Visualized Experiments). 2022;186:e64149. https://doi.org/10.3791/64149.
Leiner T, Rueckert D, Suinesiaputra A, Baeßler B, Nezafat R, Išgum I, Young AA. Machine learning in cardiovascular magnetic resonance: basic concepts and applications. J Cardiovasc Magn Reson. 2019;21(1):61. https://doi.org/10.1186/s12968-019-0575-y.
Fotaki A, Puyol-Antón E, Chiribiri A, Botnar R, Pushparajah K, Prieto C. Artificial intelligence in cardiac MRI: is clinical adoption forthcoming? Front Cardiovasc Med. 2022;8:818765. https://doi.org/10.3389/fcvm.2021.818765.
Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, Yang G-Z. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21(1):4–21. https://doi.org/10.1109/JBHI.2016.2636665.
Baeßler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54(4):221–8. https://doi.org/10.1097/RLI.0000000000000530.
Neisius U, El-Rewaidy H, Nakamori S, Rodriguez J, Manning WJ, Nezafat R. Radiomic analysis of myocardial native T1 imaging discriminates between hypertensive heart disease and hypertrophic cardiomyopathy. JACC: Cardiovasc Imaging. 2019;12(10):1946–54.
Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK,...& American Heart Association Writing Group on Myocardial Segmentation and Registration for Cardiac Imaging. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. Circulation, 2022;105(4), 539-542
Kraskov A, Stögbauer H, Grassberger P. Estimating mutual information. Phys Rev E. 2004;69(6):066138. https://doi.org/10.1103/PhysRevE.69.066138.
Chen YW, & Lin CJ (2006) Combining SVMs with various feature selection strategies. In I. Guyon, M. Nikravesh, S. Gunn, & L. A. Zadeh (Eds.), Feature Extraction (Vol. 207, pp. 315–324). Berlin, Heidelberg: Springer Berlin Heidelberg. https://doi.org/10.1007/978-3-540-35488-8_13
Spencer R, Thabtah F, Abdelhamid N, Thompson M. Exploring feature selection and classification methods for predicting heart disease. Digital Health. 2020;6:2055207620914777. https://doi.org/10.1177/2055207620914777.
Pal M, Parija S. Prediction of heart diseases using random forest. J Phys: Conf Ser. 2021;1817(1):012009. https://doi.org/10.1088/1742-6596/1817/1/012009.
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, van Mulbregt P. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods. 2020;17(3):261–72. https://doi.org/10.1038/s41592-019-0686-2.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O,…Cournapeau D. (n.d.) Scikit-learn: machine learning in Python. MACHINE LEARNING IN PYTHON.
Hillier E, Plasa G, Luu J, Benovoy M, Friedrich M G. Machine learning analysis of oxygenation-sensitive cardiovascular magnetic resonance imaging in patients with coronary artery stenosis. Society of Cardiovascular Magnetic Resonance Imaging. 24–26. San Diego, California. USA: Poster Presentation; 2023.
Ghantous CM, Kamareddine L, Farhat R, Zouein FA, Mondello S, Kobeissy F, Zeidan A. Advances in cardiovascular biomarker discovery. Biomedicines. 2020;8(12):552. https://doi.org/10.3390/biomedicines8120552.
Hathaway QA, Roth SM, Pinti MV, Sprando DC, Kunovac A, Durr AJ,…Hollander JM (2019) Machine-learning to stratify diabetic patients using novel cardiac biomarkers and integrative genomics. Cardiovasc Diabetol, 18(1), 78. https://doi.org/10.1186/s12933-019-0879-0
Jack CR, Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB,…Dubois B (2016) A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology, 87(5), 539–547. https://doi.org/10.1212/WNL.0000000000002923
Funding
Parts of the work were funded by the MUHC McGill University Health Center Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5).
Consent to Participate
Informed consent was obtained from all patients for being included in the study.
Competing Interests
The authors declare no competing interests.
Additional information
Editor-in-Chief Enrique Lara-Pezzi oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Plasa, G., Hillier, E., Luu, J. et al. Automated Data Transformation and Feature Extraction for Oxygenation-Sensitive Cardiovascular Magnetic Resonance Images. J. of Cardiovasc. Trans. Res. (2024). https://doi.org/10.1007/s12265-023-10474-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12265-023-10474-7