Abstract
Insulin resistance in skeletal muscle is an early phenomenon in the pathogenesis of type 2 diabetes. Muscle is mainly responsible for insulin-stimulated glucose clearance from the bloodstream. Thus, regulation of gene expression in muscle tissue may be involved in the pathogenesis of insulin resistance. The objective was to investigate gene expression and metabolic pathways alterations in skeletal muscle tissue following an euglycemic-hyperinsulinemic clamp in obese insulin-resistant subjects. We carried out a transcriptome comparison of skeletal muscle tissue before and after a 3-h euglycemic-hyperinsulinemic clamp following 8-week supplementation with n-3 polyunsaturated fatty acid (PUFA) (1.8 g/day) with or without a supplement of fish gelatin (FG) (25 % of daily protein intake) in 16 obese insulin-resistant subjects. Results indicate that approximately 5 % (1932) of expressed transcripts were significantly changed after the clamp in both n-3 PUFA and n-3 PUFA + FG supplementation periods. Of these differentially expressed transcripts, 1394 genes associated with enzymes, transcription and translation regulators, transporters, G protein-coupled receptors, cytokines, and ligand-dependent nuclear receptors were modified. Metabolic pathways that were significantly modified included liver X receptor/retinoid X receptors (RXR) activation, vitamin D receptor/RXR activation, interleukin (IL)-8, acute phase response, IL10, triggering receptor expressed on myeloid cells 1, peroxisome proliferator-activated receptor, G-beta/gamma and hepatocyte growth factor and IL6 signaling. Taken together, results suggest that mainly inflammatory and transcription factors are modified following clamp in obese insulin-resistant subjects. Overall, understanding the changes in metabolic pathways due to insulin may be a potential target for the management of insulin resistance.
Similar content being viewed by others
Introduction
Insulin resistance is the main metabolic feature of type 2 diabetes, and several studies indicate that it generally precedes the onset of the disease (Eriksson et al. 1989; Martin et al. 1992). Despite the severe impairment in insulin action in insulin-resistant states, glucose tolerance remains normal because the pancreatic beta cells are able to augment their insulin secretory capacity to offset the insulin resistance. Hyperinsulinemia occurs as a result of the increase in insulin secretion from the beta cells. Over time, the excessive rates of insulin secretion cannot be maintained, leading to the development of glucose intolerance and type 2 diabetes.
Skeletal muscle is the major site for insulin-dependent glucose disposal, and type 2 diabetic patients are characterized by a marked decrease in insulin-stimulated glucose utilization in muscle mainly due to reduced glucose uptake and storage (Ducluzeau et al. 2001; Yki-Jarvinen 1995). Skeletal muscle insulin resistance is critical to the pathogenesis of type 2 diabetes and the metabolic syndrome. Consequently, gene expression studies aimed at identifying key genes that might be responsible for insulin resistance and type 2 diabetes focused on muscle tissue. Previously, in vivo studies suggest that gene expression regulating oxidative phosphorylation and aerobic capacity are reduced in skeletal muscle of patients with type 2 diabetes compared with normal patients (Mootha et al. 2003; Patti et al. 2003). Taken together, gene expression profiles in insulin-resistant and diabetics subjects are different from healthy individuals.
Microarray analysis offers a powerful tool for characterizing large-scale changes in gene expression levels. However, the large number of genes tested and high variability between individuals and limited sample sizes in human studies make it difficult to distinguish true differences from noise when alterations in gene expression are modest. It is well accepted that the hyperinsulinemic clamp method allows the individual effect of insulin to be studied (Rome et al. 2003). The main objective of the current study is to explore the changes in gene expression levels associated with glucose, fatty acid, and inflammation biological pathways, related to the effect of insulin injected during a hyperinsulinemic-euglycemic clamp in insulin-resistant obese subjects. In order to do so, we analyzed the commonly differentiated gene expression after two distinct supplementations including n-3 polyunsaturated fatty acid (PUFA) and n-3 PUFA + fish gelatin (+FG), which allowed us to examine solely the effects of the hyperinsulinemic-euglycemic clamp and to avoid confounding factors due to the diets. In addition, using biological pathways approach among genes, rather than individual genes, may allow us to detect more subtle changes in metabolic pathways.
Materials and methods
Experimental design
Twenty-one males, as well as pre- and postmenopausal female participants aged 35–70 years were recruited in the Quebec City greater area. Participants had a body mass index (BMI) over 25 kg/m2 and had a fasting plasma insulin >90 pmol/L including a fasting plasma glucose <7.0 mmol/L and 2-h plasma glucose <11.1 mmol/L. Subjects were excluded from the study if they had weight change ±10 % within the 6 months prior to study onset, a major surgery in the 3 months prior to study onset, diabetes, familial or primary hyperlipidemia, hepatic or metabolic diseases, smoking, chronic hypertension (>160/100 mmHg), incompatibility with fish consumption (allergy, intolerance, or dislike), and medications known to affect lipid and glucose metabolism. In order to reduce metabolic variations, the tests were performed in the follicular phase for pre-menopausal women. Fasting blood samples were collected for plasma biochemistry and hematology. The Clinical Research Ethical Committee of Laval University Hospital Center and the Food Directorate of Health Canada approved the experimental protocol. Prior to the study entry, participants received a complete description of the protocol, and their written informed consent was obtained. This trial was registered at clinical trials gov as NCT01215903.
A clinical trial was carried out at the Institute of Nutraceuticals and Functional Foods (INAF) center of Laval University. This sub-study was part of a larger study that was previously described (Rudkowska et al. 2011). Briefly, individual dietary instructions were given by a trained dietician to achieve the National Cholesterol Education Program (NCEP) Step 1 diet guidelines (2001). Subjects were asked to follow these dietary recommendations and maintain their body weight stable throughout the protocol. After a 4-week run-in where the subjects were not allowed to consume any fish products, each participant was randomized to consume with the NCEP Step 1 diet either the n-3 PUFA supplementation or the n-3 PUFA + FG supplementation for 8 weeks each. Participants switched back to their usual diet for a washout period of 8 weeks including a 4-week run-in period. Then, they crossed over to the other experimental treatment for an additional 8 weeks.
For the n-3 PUFA supplementation, each subject received a bottle containing all needed fish oil capsules for the 8 weeks, which provided the equivalent of a total of 1.8 g of EPA and DHA daily. For the n-3 PUFA + FG supplementation, each subject received a bottle containing all needed fish oil capsules for the 8 weeks, which provided the equivalent of a total of 1.8 g of EPA and DHA daily, and FG incorporated into a broth, which provided approximately 25 % of their daily protein intake. Compliance was assessed from the return of capsules of n-3 PUFA and broth cube supplements. Subjects were asked to report any deviation during the protocol.
Subjects were submitted to a 3-h euglycemic-hyperinsulinemic clamp as previously described by (Defronzo et al. 1979). Identical in vivo experiments were performed after each supplementation period.
Samples
The muscle biopsy was performed after each 8-week supplementation (n-3 PUFA and n-3 PUFA + FG) pre- and post-euglycemic-hyperinsulinemic clamp. Biopsies (approximately 50 mg) were obtained from the vastus lateralis muscle under local anesthesia using a needle with suction applied. Total RNA was extracted using the RNeasy Fibrous Tissue Mini Kit (QIAGEN, Mississauga, ON, Canada) according to the manufacturer’s instructions and stored at −80 °C. After spectrophotometric quantification and verification of the total RNA quality via the Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, United States), samples were sent for microarray analysis. Samples were excluded from additional analysis on microarray chips if they had poor RNA quality [RNA integrity number (RIN) < 8].
Array, hybridizations, and measurements
200 ng of total RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification kit (Ambion). cRNA quality was assessed by capillary electrophoresis on Agilent 2100 Bioanalyzer. Expression levels of 48,803 mRNA transcripts, to investigate 37,804 genes, were assessed by the Human-6 v3 Expression BeadChips (Illumina). Hybridization and measurements were carried out according to the manufacturer’s instructions at the McGill University/Génome Québec Innovation Center (Montreal, Canada).
Normalization controls
This article considered the minimum information about a microarray experiment (MIAME) which describes the minimum information required to ensure that microarray data can be easily interpreted and that results derived from its analysis can be independently verified (Brazma et al. 2001). First, filtering was done with the expression data of the lowest detection microarray score in each tissue type. A detection score on microarrays of a P value threshold of 0.01 was used to define the number of transcripts detected, as recommended by manufacturer and implemented in FlexArray software (version 1.4.1) (Blazejczyk and Nadon 2007). Secondly, the Lumi algorithm was used to normalize Illumina microarray data. Specifically, expression values were normalized by using Lumi via the robust multi array average (RAM) algorithm (Bolstad et al. 2003). This step was followed by quantile normalization and log2 transformation. The samples were then grouped according to diet (n-3 PUFA or n-3 PUFA + FG) and time (pre- and post-euglycemic-hyperinsulinemic clamp). To assess the effect of insulin on gene expression, we performed a significance analysis of microarrays (SAM) algorithm, an adaptation of a t test between the basal state (pre-clamp) and the insulin-stimulated state (post-clamp) for microarray data, on all probes. In general, the SAM application assigns a score to gene on basis of change in gene expression relative to standard deviation of repeated measurements. Then, SAM uses permutations of the repeated measurements to estimate the false discovery rate (Tusher et al. 2001). A cutoff of P ≤ 0.05 was used to select the regulated genes. In addition, a fold change cutoff was also computed by FlexArray software to assess the level and the direction of the gene regulation. This fold change is calculated as the absolute ratio of normalized intensities between the mean values of all individual FC (post-/pre-clamp). Thus, two cutoff values were used to minimize the chances of false positives. Fold changes at >2 and P ≤ 0.05 (up-regulated) or fold changes at <0.5 and P ≤ 0.05 (down-regulated) were taken from each treatment to determine differentially expressed transcripts, and transcript lists were generated.
Biological pathway analyses
Lists of differentially expressed genes were imported into Ingenuity Pathway Analysis (IPA) system (Ingenuity® Systems, www.ingenuity.com). Pathway analyses allowed to determine whether genes found to be differentially expressed belong to pre-defined networks more than expected by chance alone and help to add structure to the vast amount of data generated by microarrays. The IPA system was used to visualize gene expression data in the context of biological pathways. First, two input files were uploaded in the IPA system: (1) fold changes of all probe sets between pre- and post-euglycemic-hyperinsulinemic clamp following an n-3 PUFA supplementation and (2) fold changes of all probe sets between pre- and post-euglycemic-hyperinsulinemic clamp following n-3 PUFA + FG supplementation, and dataset in core analysis was created. Further, the core dataset was analyzed using the general settings for IPA system as “ingenuity knowledge base (genes)” and “considered only molecules and/or relationships where species is humans”. As the reference set, the ingenuity knowledge base (genes + endogenous chemicals) was used. Finally, the canonical pathways analysis was used to identify the most significant pathways.
Real-time RT-PCR
cDNA was mixed with TaqMan Universal PCR Master Mix (Applied Biosystems) and a gene-specific primer and probe mixture (predeveloped TaqMan gene expression assays; Applied Biosystems) in a final volume of 20 μL. The assays used were as follows: Hs00174103_m1 (IL8), Hs00264920_s1 (THBD), Hs00231069_m1 (ATF3), and GAPDH as the housekeeping gene Hs99999905_m1 (GAPDH). All assays used the same fluorescent reporter probe (FAM™ dye-labeled), and thus, each combination treatment and gene was analyzed in individual wells on a 96-well plate. All samples were run in duplicate on an Applied Biosystem 7500 fast real-time PCR system (Applied Biosystems) using the following thermal cycling profile: 50 °C (2 min), 95 °C (10 min), followed by 40 steps of 95 °C for 15 s, and 60 °C for 60 s. The RT-PCR results were imported into Microsoft Excel, and the average value of duplicate C t values calculated. Data were adjusted for the endogenous control (GAPDH). Data were analyzed using a paired t test to determine significant changes between pre- and post-euglycemic-hyperinsulinemic clamp. Pearson correlations were performed between RT-PCR and microarray data. Statistical significance was defined as P ≤ 0.05.
Results
Characteristics of the subjects and effects of supplementations
From the 21 subjects who started the study, sixteen subjects, including 7 men and 9 women, aged 57 ± 2 year, and with BMI of 30 ± 1.0 kg/m2, completed the study. Five subjects became diabetic and were no longer eligible for this study. Expectedly, the 16 subjects had sub-optimal glycemic control: fasting plasma glucose of 5.98 ± 0.08 mmol/L, 2-h plasma glucose of 7.38 ± 0.40 mmol/L, and fasting plasma insulin of 106 ± 9 ρmol/L (mean ± SEM). In addition, subjects had slightly deteriorated plasma lipid levels: total cholesterol (TC): 5.51 ± 0.30 mmol/L, LDL-C: 3.84 ± 0.25 mmol/L, HDL-C: 1.02 ± 0.05 mmol/L, and TG: 1.51 ± 0.11 mmol/L.
Changes in gene expression levels
RNA extraction from skeletal muscle tissues was performed on samples: 16 pre-clamp and 16 post-clamp after each supplementation periods (total of 64 skeletal muscle tissues samples). After verification of the total RNA quality, 4 samples were excluded from further analysis on microarray chips because of poor RNA quality (RIN < 8). The final analysis consisted of 60 samples taken at 4 time points: pre-clamp n-3 PUFA supplementation (n = 16), pre-clamp n-3 PUFA + FG (n = 13), post-clamp n-3 PUFA supplementation (n = 16), and post-clamp n-3 PUFA + FG (n = 15). Consequently, microarray analysis was performed on 60 samples to gain insight on changes in gene expression levels following a hyperinsulinemic-euglycemic clamp.
First, results demonstrate that of the 48,803 transcripts present, approximately 36,738 (75 %) were detected in skeletal muscle cells using the detection P value threshold of 0.01. Therefore, approximately 24,000 genes were expressed in skeletal muscle cells.
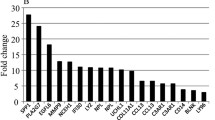
Secondly, we compared changes in gene expression levels of samples taken pre- and post-clamp on the n-3 PUFA supplementation period. Results demonstrate that 2749 transcripts were differentially expressed (70 % (1920) down-regulated and 30 % (829) up-regulated) (Fig. 1) between the pre- and post-clamp intervention following the n-3 PUFA supplementation period. Of the 2749 differentially expressed transcripts, 1874 genes were associated with a mapped gene by the IPA system. Further, we analyzed the results of gene expression levels pre- to post-clamp after the n-3 PUFA + FG intervention. Results reveal that 2452 transcripts were differentially expressed (67 % (1644) down-regulated and 33 % (808) up-regulated) (Fig. 1). Of the 2452 differentially expressed transcripts, 1785 genes were associated with a mapped gene by the IPA system.
Finally, we compared the results of the changes in gene expression between both supplementation periods; this allowed us to delete the effects of the diets/supplementation/external factors and examine solely the effects of the hyperinsulinemic-euglycemic clamp. There were 1932 transcripts that were commonly changed in both n-3 PUFA and n-3 PUFA + FG supplementations periods (Fig. 1). Of these 1932 commonly differentially expressed transcripts, 1394 genes were associated with a mapped gene by the IPA system. These genes were grouped: enzymes (188), transcription and translation regulators (132), transporter (67), G protein-coupled receptors (35), cytokines (25), ligand-dependent nuclear receptors (6) and others (741). In addition, the up-regulated were similar after both supplementation periods (Table 1: Top up-regulated genes in skeletal muscle tissue following hyperinsulinemic-euglycemic clamp.) Further, the top down-regulated genes were regulated to a lower magnitude [−7.65- to −4.6-fold change (max to min)] compared with the top up-regulated genes [+191.44- to 17.63-fold change (max to min)] (data not shown).
Pathway analysis results
IPA indicates that the following pathways were changed following clamp to similar extents after the n-3 PUFA and the n-3 PUFA + FG supplementation: liver X receptor/retinoid X receptors (RXR) activation, vitamin D receptor (VDR)/RXR activation, interleukin (IL)-8 signaling, acute phase response signaling, IL-10 signaling, triggering receptor expressed on myeloid cells 1 (TREM1) signaling, peroxisome proliferator-activated receptor (PPAR) signaling, G-beta gamma signaling, and hepatocyte growth factor (HGF) signaling and IL-6 signaling (Fig. 2).
Results for real-time RT-PCR
Results from RT-PCR demonstrate that pre- to post-clamp results were differentially expressed by 1.47 fold change for IL8 (P < 0.001), 2.29 fold change for THBD (P < 0.001), and 1.94 fold change for ATF3 (P < 0.001) from pre-clamp results after n-3 PUFA and n-3 PUFA + FG supplementation (no significant differences between diets). In addition, expression levels obtained by RT-PCR and microarray were highly correlated for IL8 (P < 0.001), THBD (P < 0.001), and ATF3 (P < 0.001). In addition, we can confirm that the kinetics of insulin-induced expression patterns by RT-PCR methods are comparable with microarray results.
Discussion
Approximately 5 % (1932) of expressed transcripts were significantly changed by insulin in obese insulin-resistant subjects. Metabolic pathway analyses indicate generally changes in pro-inflammatory, transcriptional, and G protein-coupled receptor (GPCR) signaling pathways. Previous studies have examined the rapid changes in the gene expression levels in skeletal muscle during euglycemic-hyperinsulinemic clamp conditions in healthy subjects (Coletta et al. 2008; Rome et al. 2003; Wu et al. 2007). These studies demonstrate changes in gene expression levels of genes related mainly to transcription factors, intermediary and energy metabolisms, immune response, and intracellular signaling in healthy volunteers (Coletta et al. 2008; Rome et al. 2003; Wu et al. 2007). Additionally, a study demonstrated that the beginning of insulin stimulation changes are usually apparent in the expression of early response transcription factors together with early signaling molecules, chemokines, and receptors followed afterward by secreted growth factors and proteases, metabolic enzymes and cell-cycle-regulating genes (Hansen et al. 2004). Thus, the limited stability of these genes categories may facilitate rapid regulatory control for intracellular signaling to reflect the current condition of an individual. The current results concur with previous investigations of the effect of insulin on the transcriptional response factors; however, a larger proportion of changed transcripts were related to inflammation signaling in the obese insulin-resistant subjects.
Insulin resistance is associated with increased levels of inflammation markers (Garcia et al. 2010). Moreover, it is well-know that cross-talks occur between insulin and cytokine signaling pathways. Results of the present study demonstrate that a large number of inflammatory cytokines—IL8, IL6, CCL2, CCL8, and CXCL2—were up-regulated by insulin. Additionally, GADD45B, THBD, and SOCS3 that are tightly regulated to inflammatory response, were also strongly up-regulated after insulin challenge. Previous studies have demonstrated that these genes are strongly up-regulated after a clamp challenge (Hansen et al. 2004; Rome et al. 2003). These genes changed various pathways related to inflammatory processes: IL8, acute phase response, IL10, TREM1 and IL6 signaling. Overall, these results are consistent with those of previous in vitro studies (Hansen et al. 2004), which have shown that insulin induces a pro-inflammatory immune responses following insulin challenge.
Earlier studies have shown that in healthy subjects insulin increased a number of inflammatory genes (Coletta et al. 2008; Rome et al. 2003); however, to a lesser extent than the current study. This difference in magnitude may be due to the insulin sensitivity, obesity status of subjects, as well as the subjects’ ethnic origins. First, a recent study showed that the expression of key genes in the inflammatory pathways was higher in the insulin-resistant than in the insulin-sensitive groups (Westerbacka et al. 2006). Secondly, a chronic systemic low-grade inflammation is typically observed in obese subjects. The beneficial effect of weight loss was associated with the modification of the inflammation-related gene expression (Clement et al. 2004; de Mello et al. 2008). Further, gene expression in obese subjects after dietary restriction was closer to the profile of lean subjects than to the pattern of obese subjects before dietary restriction (Clement et al. 2004). Finally, researchers have compared the effect of a clamp between ethnicities; they found that the inflammatory genes were increased to a greater level in European-American compared with African-Americans subjects (Elbein et al. 2011; Smith et al. 2010). These results suggest that the expression levels of inflammatory genes is greater in our population of French-Canadian, obese, insulin-resistant subjects’ compared with previous populations.
The expression of transcription regulation factors, such as FOS, FOSB, JUNB, EGR1, EGR2, IER3, MYC, CDKN1A, and BCL3 were also altered by insulin. These genes control a number of cellular processes including differentiation, proliferation, and apoptosis. Further, the LXR/RXRs, PPAR, VDR-RXR, and HGF signaling pathways were modified. These pathways involve nuclear receptors that mediate the transcriptional regulation of a number of genes. It is well known that insulin increases fatty acid uptake and triglyceride storage in skeletal muscle. LXRs control cholesterol homeostasis in the body as well as synthesis of fatty acids and triglycerides, which are secreted and used for the formation of lipoproteins (Repa and Mangelsdorf 2002). A study has demonstrated that LXRs may also play a role in glucose homeostasis (Cao et al. 2003). Further, PPARs are nuclear receptors that are activated by fatty acids and their derivatives. PPARA is highly expressed in skeletal muscle and the liver, and it plays a role in the clearance of cellular lipids via the regulation of genes involved in lipid metabolism as well as cytokine production (Alwayn et al. 2006; Israelian-Konaraki and Reaven 2005). PPARB/D is involved in lipid oxidation and cell proliferation. PPARG is expressed mainly in fat but also to a lesser extent in muscle and is important for adipogenesis, lipid metabolism, inflammation, and glucose control (Yki-Jarvinen 1995; Zierath et al. 1998). Further, VDR/RXR signaling regulates a variety of metabolic pathways together with a variety of immune responses (Bouillon et al. 2008). In addition, an animal study showed that mice lacking a functional VDR had reduced insulin secretion and higher blood glucose levels than normal mice (Zeitz et al. 2003). Therefore, VDR/RXR signaling may be an important target for glycemic control. The HGF is a multi-potent cytokine that stimulates cell proliferation, differentiation, motility, and invasiveness. Previous studies have reported similar findings concerning the effect of insulin on the expression of transcription factors (Hansen et al. 2004; Rome et al. 2003). Overall, these above transcription factors signaling pathways are essential for the energy metabolism and intracellular signaling pathways to adapt to environmental changes, such as insulin, and may also participate in the pathogenesis of the disease (Barroso et al. 1999; Brunetti et al. 1996).
GPCR pathways control glucose and fatty acid metabolism and the onset of obesity and diabetes. The GPCR pathways involve two principal signaling pathways: the cAMP and the phosphatidylinositol signal pathway. In the current study, the regulator of G protein signaling 16 (RGS16) was one of the most up-regulated gene after the clamp. A previous study demonstrated that RGS16 gene provides a signaling mechanism for glucose production to inhibit GPCR-stimulated fatty acid oxidation in hepatocytes (Pashkov et al. 2011). In accordance, the G-beta/gamma signaling pathway was strongly modified after the clamp challenge. Overall, insulin significantly influences GPCR pathways.
In conclusion, these results illustrate the marked effect of insulin on pro-inflammatory, transcription, and GPCR pathways in obese Caucasian insulin-resistant subjects. Previously Wu et al. (2007) demonstrated that the effects of a euglycemic-hyperinsulinemic clamp were noted on all stages including transcription, mRNA processing, translation, post-translational protein modification, and protein degradation. In sum, these results suggest that key inflammatory and transcriptional pathways should be targeted in the development of nutritional as well as pharmaceutical therapies to decrease the risk of progression to type 2 diabetes for obese insulin-resistant subjects.
References
Alwayn IP, Andersson C, Lee S, Arsenault DA, Bistrian BR, Gura KM, Nose V, Zauscher B, Moses M, Puder M (2006) Inhibition of matrix metalloproteinases increases PPAR-alpha and IL-6 and prevents dietary-induced hepatic steatosis and injury in a murine model. Am J Physiol Gastrointest Liver Physiol 291:G1011–G1019
Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O’Rahilly S (1999) Dominant negative mutations in human PPAR gamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883
Blazejczyk MironM, Nadon R (2007) FlexArray: a statistical data analysis software for gene expression microarrays. Genome Quebec, Montreal
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Bouillon R, Carmeliet G, Verlinden L, van EE, Verstuyf A, Luderer HF, Lieben L, Mathieu C, Demay M (2008) Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev 29:726–776
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29:365–371
Brunetti A, Brunetti L, Foti D, Accili D, Goldfine ID (1996) Human diabetes associated with defects in nuclear regulatory proteins for the insulin receptor gene. J Clin Invest 97:258–262
Cao G, Liang Y, Broderick CL, Oldham BA, Beyer TP, Schmidt RJ, Zhang Y, Stayrook KR, Suen C, Otto KA, Miller AR, Dai J, Foxworthy P, Gao H, Ryan TP, Jiang XC, Burris TP, Eacho PI, Etgen GJ (2003) Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J Biol Chem 278:1131–1136
Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, Zucker JD, Vidal H, Laville M, Barsh GS, Basdevant A, Stich V, Cancello R, Langin D (2004) Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J 18:1657–1669
Coletta DK, Balas B, Chavez AO, Baig M, Abdul-Ghani M, Kashyap SR, Folli F, Tripathy D, Mandarino LJ, Cornell JE, Defronzo RA, Jenkinson CP (2008) Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am J Physiol Endocrinol Metab 294:E910–E917
de Mello VD, Kolehmainen M, Pulkkinen L, Schwab U, Mager U, Laaksonen DE, Niskanen L, Gylling H, Atalay M, Rauramaa R, Uusitupa M (2008) Downregulation of genes involved in NFkappaB activation in peripheral blood mononuclear cells after weight loss is associated with the improvement of insulin sensitivity in individuals with the metabolic syndrome: the GENOBIN study. Diabetologia 51:2060–2067
Defronzo RA, Tobin JD, Andres R (1979) Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol 237:E214–E223
Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, Riou JP, Vidal H (2001) Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes 50:1134–1142
Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK (2011) Global Gene expression profiles of subcutaneous adipose and muscle from Glucose-Tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes 60:1019–1029
Eriksson J, Franssila-Kallunki A, Ekstrand A, Saloranta C, Widen E, Schalin C, Groop L (1989) Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N Engl J Med 321:337–343
Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) (2001) Expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III). JAMA 285:2486–2497
Garcia C, Feve B, Ferre P, Halimi S, Baizri H, Bordier L, Guiu G, Dupuy O, Bauduceau B, Mayaudon H (2010) Diabetes and inflammation: fundamental aspects and clinical implications. Diabetes Metab 36:327–338
Hansen L, Gaster M, Oakeley EJ, Brusgaard K, Damsgaard Nielsen EM, Beck-Nielsen H, Pedersen O, Hemmings BA (2004) Expression profiling of insulin action in human myotubes: induction of inflammatory and pro-angiogenic pathways in relationship with glycogen synthesis and type 2 diabetes. Biochem Biophys Res Commun 323:685–695
Israelian-Konaraki Z, Reaven PD (2005) Peroxisome proliferator-activated receptor-alpha and atherosclerosis: from basic mechanisms to clinical implications. Cardiol Rev 13:240–246
Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR (1992) Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet 340:925–929
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273
Pashkov V, Huang J, Parameswara VK, Kedzierski W, Kurrasch DM, Tall GG, Esser V, Gerard RD, Uyeda K, Towle HC, Wilkie TM (2011) Regulator of G protein signalling (RGS16) inhibits hepatic fatty acid oxidation in a carbohydrate response element-binding protein (ChREBP)-dependent manner. J Biol Chem 286:15116–15125
Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100:8466–8471
Repa JJ, Mangelsdorf DJ (2002) The liver X receptor gene team: potential new players in atherosclerosis. Nat Med 8:1243–1248
Rome S, Clement K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS, Riou JP, Laville M, Vidal H (2003) Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperinsulinemic clamp. J Biol Chem 278:18063–18068
Rudkowska I, Ponton A, Jacques H, Lavigne C, Holub BJ, Marette A, Vohl MC (2011) Effects of a supplementation of n-3 polyunsaturated fatty acids with or without fish gelatin on gene expression in peripheral blood mononuclear cells in obese, insulin-resistant subjects. J Nutrigenet Nutrigenomics 4:192–202
Smith LM, Yao-Borengasser A, Starks T, Tripputi M, Kern PA, Rasouli N (2010) Insulin resistance in African-American and Caucasian women: differences in lipotoxicity, adipokines, and gene expression in adipose tissue and muscle. J Clin Endocrinol Metab 95:4441–4448
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121
Westerbacka J, Corner A, Kannisto K, Kolak M, Makkonen J, Korsheninnikova E, Nyman T, Hamsten A, Fisher RM, Yki-Jarvinen H (2006) Acute in vivo effects of insulin on gene expression in adipose tissue in insulin-resistant and insulin-sensitive subjects. Diabetologia 49:132–140
Wu X, Wang J, Cui X, Maianu L, Rhees B, Rosinski J, So WV, Willi SM, Osier MV, Hill HS, Page GP, Allison DB, Martin M, Garvey WT (2007) The effect of insulin on expression of genes and biochemical pathways in human skeletal muscle. Endocrine 31:5–17
Yki-Jarvinen H (1995) Role of insulin resistance in the pathogenesis of NIDDM. Diabetologia 38:1378–1388
Zeitz U, Weber K, Soegiarto DW, Wolf E, Balling R, Erben RG (2003) Impaired insulin secretory capacity in mice lacking a functional vitamin D receptor. FASEB J 17:509–511
Zierath JR, Ryder JW, Doebber T, Woods J, Wu M, Ventre J, Li Z, McCrary C, Berger J, Zhang B, Moller DE (1998) Role of skeletal muscle in thiazolidinedione insulin sensitizer (PPAR gamma agonist) action. Endocrinology 139:5034–5041
Acknowledgments
We express our gratitude to the subjects for their excellent collaboration. The work of Julie Bisson, Éliane Picard-Deland, and Julie Marois in the coordination of the clinical trial is acknowledged. We thank Catherine Raymond for contributing to the laboratory work. We also acknowledge the contribution of André Ponton and the platform technicians at McGill University/Génome Québec Innovation Center (Montreal, Canada) for microarray analysis. This work was supported by a grant from the Advanced Foods and Materials Network (AFMNet). Iwona Rudkowska is supported by a CIHR Bisby Postdoctoral Fellowship Award (200810BFE). André Marette holds a CIHR/Pfizer Research Chair on the pathogenesis of insulin resistance and cardiovascular disease. Marie-Claude Vohl holds a Tier 1 Canada Research Chair in Genomics Applied to Nutrition and Health.
Conflict of interest
There are no conflicts of interest to be declared.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rudkowska, I., Jacques, H., Weisnagel, S.J. et al. Transcriptomic profiles of skeletal muscle tissue following an euglycemic-hyperinsulinemic clamp in insulin-resistant obese subjects. Genes Nutr 8, 91–98 (2013). https://doi.org/10.1007/s12263-012-0298-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12263-012-0298-2