Abstract
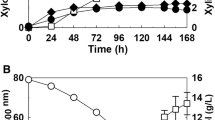
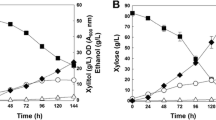
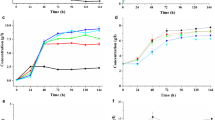
Xylose fermentation has been reported to be improved in Kluyveromyces marxianus via strain improvement by overexpressing xylose reductase (XYL1) and xylitol dehydrogenase (XYL2). This study performed directed evolution to further enhance xylose consumption in a K. marxianus mutant following transcriptomic analysis to determine genes associated with enhanced characteristics. KmXYL1 and KmXYL2 genes were overexpressed in K. marxianus 17555ΔURA3 for improving xylose fermentation. By performing directed evolution, a mutant K. marxianus KHM89 showing enhanced ethanol production was isolated from xylose medium. K. marxianus KHM89 consumed 47.39 g/L of xylose and produced 22.62 g/L of xylitol and 10.59 g/L of ethanol while the parental strain consumed 25.15 g/L of xylose and produced 7.36 g/L of xylitol and 2.05 g/L of ethanol. RNA sequencing-based transcriptomic analysis showed that alcohol dehydrogenases, aldehyde dehydrogenases, and NAD+ salvage pathway enzymes were upregulated in K. marxianus KHM89. These results were achieved via a combinatorial approach of rational design and directed evolution. The findings of this study contribute to the improvement of xylose fermentation by K. marxianus at an industrial scale.
Similar content being viewed by others
References
Lee, R. A. and J.-M. Lavoie (2013) From first- to third-generation biofuels: challenges of producing a commodity from a biomass of increasing complexity. Anim. Front. 3: 6–11.
Hamelinck, C. N., G. van Hooijdonk, and A. P. Faaij (2005) Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle- and long-term. Biomass Bioenergy. 28: 384–410.
Sun, Y. and J. Cheng (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour. Technol. 83: 1–11.
Balat, M. (2009) Bioethanol as a vehicular fuel: a critical review. Energy Sources Part A. 31: 1242–1255.
Saini, J. K., R. Saini, and L. Tewari (2015) Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments. 3 Biotech. 5: 337–353.
Banerjee, S., G. Mishra, and A. Roy (2019) Metabolic engineering of bacteria for renewable bioethanol production from cellulosic biomass. Biotechnol. Bioprocess Eng. 24: 713–733.
Lee, J. (1997) Biological conversion of lignocellulosic biomass to ethanol. J. Biotechnol. 56: 1–24.
Sivakumar, G., D. R. Vail, J. Xu, D. M. Burner, J. O. Lay, X. Ge, and P. J. Weathers (2010) Bioethanol and biodiesel: alternative liquid fuels for future generations. Eng. Life Sci. 10: 8–18.
Demirbaş, A. (2004) Ethanol from cellulosic biomass resources. Int. J. Green Energy. 1: 79–87.
Wang, Y., X. Yu, and H. Zhao (2020) Biosystems design by directed evolution. AIChE J. 66: e16716.
Eriksen, D. T., P. C. H. Hsieh, P. Lynn, and H. Zhao (2013) Directed evolution of a cellobiose utilization pathway in Saccharomyces cerevisiae by simultaneously engineering multiple proteins. Microb. Cell Fact. 12: 61.
Crook, N., J. Abatemarco, J. Sun, J. M. Wagner, A. Schmitz, and H. S. Alper (2016) In vivo continuous evolution of genes and pathways in yeast. Nat Commun. 7: 13051.
Mans, R., J.-M. G. Daran, and J. T. Pronk (2018) Under pressure: evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol. 50: 47–56.
Kim, S.-B., D.-H. Kwon, J.-B. Park, and S.-J. Ha (2019) Alleviation of catabolite repression in Kluyveromyces marxianus: the thermotolerant SBK1 mutant simultaneously coferments glucose and xylose. Biotechnol. Biofuels. 12: 90.
Lee, S.-M., T. Jellison, and H. S. Alper (2012) Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78: 5708–5716.
Parreiras, L. S., R. J. Breuer, R. Avanasi Narasimhan, A. J. Higbee, A. La Reau, M. Tremaine, L. Qin, L. B. Willis, B. D. Bice, B. L. Bonfert, R. C. Pinhancos, A. J. Balloon, N. Uppugundla, T. Liu, C. Li, D. Tanjore, I. M. Ong, H. Li, E. L. Pohlmann, J. Serate, S. T. Withers, B. A. Simmons, D. B. Hodge, M. S. Westphall, J. J. Coon, B. E. Dale, V. Balan, D. H. Keating, Y. Zhang, R. Landick, A. P. Gasch, and T. K. Sato (2014) Engineering and two-stage evolution of a lignocellulosic hydrolysate-tolerant Saccharomyces cerevisiae strain for anaerobic fermentation of xylose from AFEX pretreated corn stover. PLoS One. 9: e107499.
Kwon, D.-H., J.-B. Park, E. Hong, and S.-J. Ha (2019) Ethanol production from xylose is highly increased by the Kluyveromyces marxianus mutant 17694-DH1. Bioprocess Biosyst. Eng. 42: 63–70.
Nagasaki, H., Y. Suzuki, T. Fujimoto, H. Saito, T. Suzuki, and S. Watanabe (2021) Effect of air sparging on ethanol production from xylose and glucose in continuous chemostat fermentation process utilizing high cell density of Candida intermedia 4-6-4T2. J. Jpn. Pet. Inst. 64: 178–187.
Schmidt, M. T., B. C. Smith, M. D. Jackson, and J. M. Denu (2004) Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 279: 40122–40129.
Sporty, J., S. J. Lin, M. Kato, T. Ognibene, B. Stewart, K. Turteltaub, and G. Bench (2009) Quantitation of NAD+ biosynthesis from the salvage pathway in Saccharomyces cerevisiae. Yeast. 26: 363–369.
Li, Y.-F. and W.-G. Bao (2007) Why do some yeast species require niacin for growth? Different modes of NAD synthesis. FEMS Yeast Res. 7: 657–664.
Kim, J.-S., J.-B. Park, S.-W. Jang, and S.-J. Ha (2015) Enhanced xylitol production by mutant Kluyveromyces marxianus 36907-FMEL1 due to improved xylose reductase activity. Appl. Biochem. Biotechnol. 176: 1975–1984.
Anderson, P. J., K. McNeil, and K. Watson (1986) High-efficiency carbohydrate fermentation to ethanol at temperatures above 40 degrees C by Kluyveromyces marxianus var. marxianus isolated from sugar mills. Appl. Environ. Microbiol. 51: 1314–1320.
Hughes, D. B., N. J. Tudroszen, and C. J. Moye (1984) The effect of temperature on the kinetics of ethanol production by a thermotolerant strain of Kluveromyces marxianus. Biotechnol. Lett. 6: 1–6.
Park, J.-B., J.-S. Kim, S.-W. Jang, E. Hong, and S.-J. Ha (2015) The application of thermotolerant yeast Kluyveromyces marxianus as a potential industrial workhorse for biofuel production. KSBB J. 30: 125–131.
Park, J.-B., J.-S. Kim, S.-W. Jang, D.-H. Kweon, E. K. Hong, W. C. Shin, and S.-J. Ha (2016) Sequence analysis of KmXYL1 genes and verification of thermotolerant enzymatic activities of xylose reductase from four Kluyveromyces marxianus strains. Biotechnol. Bioprocess Eng. 21: 581–586.
Park, J.-B., J.-S. Kim, D.-H. Kweon, D.-H. Kweon, J.-H. Seo, and S.-J. Ha (2019) Overexpression of endogenous xylose reductase enhanced xylitol productivity at 40 °C by thermotolerant yeast Kluyveromyces marxianus. Appl. Biochem. Biotechnol. 189: 459–470.
Jiang, M., X. Chen, L. Liang, R. Liu, Q. Wan, M. Wu, H. Zhang, J. Ma, K. Chen, and P. Ouyang (2014) Co-expression of phosphoenolpyruvate carboxykinase and nicotinic acid phosphoribosyltransferase for succinate production in engineered Escherichia coli. Enzyme Microb. Technol. 56: 8–14.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2019R1F1A1055315).
Author information
Authors and Affiliations
Contributions
Deok-Ho Kwon contributed to the study conception, design, material preparation and data collection. Suk-Jin Ha contributed to review and editing of the manuscript.
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kwon, DH., Ha, SJ. Xylose Fermentation Was Improved by Kluyveromyces marxianus KHM89 through Up-regulation of Nicotinamide Adenine Dinucleotide (NAD+) Salvage Pathway. Biotechnol Bioproc E 27, 624–630 (2022). https://doi.org/10.1007/s12257-022-0036-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-022-0036-6