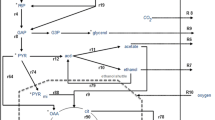
Abstract
Two novel thermotolerant yeasts, Kluyveromyces marxianus NIRE-K1.1 and Kluyveromyces marxianus NIRE-K3.1, evolutionarily adapted for fermentation of glucose and xylose were analyzed for their metabolic fluxes with an objective of maximum ethanol production. Metabolic fluxes were analyzed for these thermotolerant yeasts by co-fermenting glucose/xylose mixture in two different ratios (1:1 and 4:1). Flux balance analysis revealed the active role of pentose phosphate pathway for effective xylose utilization in both yeasts. A comparison between co-fermentation of glucose/xylose mixtures in the ratio of 1:1 and 4:1 (g/L) reveals that the flux from glucose-6-phosphate to ribulose-5-phosphate was approximately 2.56-fold and 3.75-fold higher in 1:1 mixture in K. marxianus NIRE-K1.1 and K. marxianus NIRE-K3.1, respectively. Overall, flux towards pyruvate (for ethanol production) was found to be higher in both glucose/xylose mixtures 1:1 (1.87%) and 4:1 (0.89%) for K. marxianus NIRE-K3.1 than K. marxianus NIRE-K1.1. Tricarboxylic Acid (TCA) cycle was also found to be incomplete for both the isolates which signify that most of the available substrates were utilized for ethanol production rather than biomass formation. Moreover, it was also observed that in both, the ethanol yields were found to be higher in case of K. marxianus NIRE-K3.1 than K. marxianus NIRE-K1.1, however, xylose uptake rates were higher in the later as compared to the former. Thus, this study concludes with the capable potential of both the yeasts for the production of bioethanol from glucose/xylose mixtures with higher yield and is highly correlated to the relative concentration of both xylose and glucose in a mixture.
Similar content being viewed by others
Abbreviations
- 1,3BPG:
-
1,3-Biphosphoglycerate
- 2PG:
-
2-Phosphoglycerate
- 3PG:
-
3-Phosphoglycerate
- ACCOA:
-
Acetyl CoA
- ACET:
-
Acetaldehyde
- ACT:
-
Acetate
- AKG:
-
α-ketoglutarate
- DHAP:
-
Dihydroxyacetone Phosphate
- E4P:
-
Erythrose-4-Phosphate
- EMP:
-
Embden Meyehoff Parnas
- ETH:
-
Ethanol
- F1,6BP:
-
Fructose-1,6-Bis Phosphate
- F6P:
-
Fructose-6-Phosphate
- FBA:
-
Flux Balance Analysis
- FAD:
-
Flavin Adenine Dinucleotide
- FDH:
-
Formate dehydrogenase
- G3P:
-
Glyceraldehyde-3-Phosphate
- G6P:
-
Glucose-6-Phosphate
- GLU:
-
Glucose
- GLY:
-
Glycerol
- GPD:
-
glycerol-3-phosphate dehydrogenases
- ICIT:
-
Isocitrate
- NAD:
-
Nicotinamide adenine dinucleotide
- OAA:
-
Oxaloacetate
- PEP:
-
Phosphoenol pyruvate
- PPP:
-
Pentose phosphate pathway
- PYR:
-
Pyruvate
- RIB5P:
-
Ribose-5-Phosphate
- RIBU5P:
-
Ribulose-5-Phosphate
- SED7P:
-
Sedoheptulose-7-Phosphate
- SUCC:
-
Succinate
- SUCCOA:
-
Succinyl CoA
- TCA:
-
Tricarboxylic acid
- XYL:
-
Xylose
- XYL5P:
-
Xylulose-5-Phosphate
- XYLE:
-
Xylitol (Extracellular)
- XYLI:
-
Xylitol (Intracellular)
- XYLU:
-
Xylulose
References
Arora, R., S. Behera, N. K. Sharma, and S. Kumar (2015) Bioprospecting thermostable cellulosomes for efficient biofuel production from lignocellulosic biomass. Bioresour. Bioprocess. 2: 38.
Behera, S., R. Arora, N. Nandhagopal, and S. Kumar (2014) Importance of chemical pretreatment for bioconversion of lignocellulosic biomass. Renew. Sustain. Energy Rev. 36: 91–106.
Woo, H. L., T. C. Hazen, B. A. Simmons, and K. M. De Angelis (2014) Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils. Syst Appl Microbiol. 37: 60–67.
Arora, R., S. Behera, N. K. Sharma, R. Singh, Y. K. Yadav, and S. Kumar (2014) Biochemical conversion of rice straw (Oryza sativa L.) to bioethanol using thermotolerant isolate K. marxianus NIRE-K3. Proceedings of Conference on Exploring Basic & Applied Sciences for Next Generation Frontiers. November 14–15. Punjab, India.
Arora, R., S. Behera, and S. Kumar (2015) Bioprospecting thermophilic/thermotolerant microbes for production of lignocellulosic ethanol: A future perspective. Renew. Sustain. Energy Rev. 51: 699–717.
Kumar, S., P. Dheeran, S. P. Singh, I. M. Mishra, and D. K. Adhikari (2013) Cooling system economy in ethanol production using thermotolerant yeast Kluyveromyces sp. IIPE453. Am. J. Microbiol. Res. 1: 39–44.
Kuhad, R. C., R. Gupta, Y. P. Khasa, A. Singh, and Y. H. P. Zhang (2011) Bioethanol production from pentose sugars: current status and future prospects. Renew. Sustain. Energy Rev. 15: 4950–4962.
Balat, M. (2011) Production of bioethanol from lignocellulosic materials via the biochemical pathway: A review. Energy Convers. Manag. 52: 858–875.
Wisselink, H. W., C. Cipollina, B. Oud, B. Crimi, J. J. Heijnen, J. T. Pronk, and A. J. A. van Maris (2010) Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng. 12: 537–551.
Feng, X. and H. Zhao (2013) Investigating xylose metabolism in recombinant Saccharomyces cerevisiae via 13C metabolic flux analysis. Microb. Cell Fact. 12: 114.
Kumar, S., S. P. Singh, I. M. Mishra, and D. K. Adhikari (2009) Ethanol and xylitol production from glucose and xylose at high temperature by Kluyveromyces sp. IIPE453. J. Ind. Microbiol. Biotechnol. 36: 1483–1489.
Arora, R., S. Behera, N. K. Sharma, and S. Kumar (2015) A new search for thermotolerant yeasts, its characterization and optimization using response surface methodology for ethanol production. Front. Microbiol. 6: 889.
Sharma, N. K., S. Behera, R. Arora, and S. Kumar (2016) Enhancement in xylose utilization using Kluyveromyces marxianus NIRE-K1 through evolutionary adaptation approach. Bioprocess Biosyst. Eng. 39: 835–843.
Manish, S., K. V. Venkatesh, and R. Banerjee (2007) Metabolic flux analysis of biologically hydrogen production by Escherichia coli. Int. J. Hydrogen. Energy. 32: 3820–3830.
Klapa, M. I. (2010) Metabolic flux analysis. pp. 15: 1–17. In: C. D. Smolke (ed.). The Metabolic Pathway Engineering Handbook. CRC Press, Boca Raton, FL, USA.
Chen, X., A. P. Alonso, D. K. Allen, J. L. Reed, and Y. Shachar-Hill (2011) Synergy between 13C-metabolic flux analysis and flux balance analysis for understanding metabolic adaptation to anaerobiosis in E. coli. Metab. Eng. 13: 38–48.
Edwards, J. S., M. Covert, and B. O. Palsson (2002) Metabolic modeling of microbes: the flux-balance approach. Environ. Microbiol. 4: 133–140.
Liu, Z., Y. Gao, J. Chen, T. Imanaka, J. Bao, and Q. Hua (2013) Analysis of metabolic fluxes for better understanding of mechanisms related to lipid accumulation in oleaginous yeast Trichosporon cutaneum. Bioresour. Technol. 130: 144–151.
Cheng, H. H., L. M. Whang, Q. A. Lin, I. C. Liu, and Q. W. Wu (2013) Metabolic flux network analysis of fermentative hydrogen production: Using Clostridium tyrobutyricum as an example. Bioresour. Technol. 141: 233–239.
Rafieenia, R. and S. R. Chaganti (2015) Flux balance analysis of different carbon source fermentation with hydrogen producing Clostridium butyricum using cell net analyzer. Bioresour. Technol. 175: 613–618.
Hohenschuh, W., R. Hector, and G. S. Murthy (2015) A dynamic flux balance model and bottleneck identification of glucose, xylose, xylulose co-fermentation in Saccharomyces cerevisiae. Bioresour. Technol. 188: 153–160.
Hendry, J. I., C. B. Prasannan, A. Joshi, S. Dasgupta, and P. P. Wangikar (2016) Metabolic model of Synechococcus sp. PCC 7002: prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 213: 190–197.
Zhang, L., Y. Xu, J. Gao, H. Xu, C. Cao, F. Xue, G. Ding, and Y. Peng (2016) Introduction of the exogenous NADH coenzyme regeneration system and its influence on intracellular metabolic flux of Paenibacilluspolymyxa. Bioresour. Technol. 201: 319–328.
Niu, K., X. Zhang, W. S. Tan, and M. L. Zhu (2011) Effect of culture conditions on producing and uptake hydrogen flux of biohydrogen fermentation by metabolic flux analysis method. Bioresour. Technol. 102: 7294–7300.
Zhao, H., Y. Lu, L. Wang, C. Zhang, C. Yang, and X. Xing (2015) Disruption of lactate dehydrogenase and alcohol dehydrogenase for increased hydrogen production and its effect on metabolic flux in Enterobacter aerogenes. Bioresour. Technol. 194: 99–107.
Salvetti, E., M. Fondi, R. Fani, S. Torriani, and G. E. Felis (2013) Evolution of lactic acid bacteria in the order Lactobacillales as depicted by analysis of glycolysis and pentose phosphate pathways. Syst. Appl. Microbiol. 36: 291–305.
Granstrom, T., A. A. Aristidou, and M. Leisola (2002) Metabolic flux analysis of Candida tropicalis growing on xylose in an oxygen-limited chemostat. Metab. Eng. 4: 248–256.
Cakir, T., K. Y. Arga, M. M. Altintas, and K. O. Ulgen (2004) Flux analysis of recombinant Saccharomyces cerevisiae YPB-G utilizing starch for optimal ethanol production. Process Biochem. 39: 2097–2108.
Sharma, N. K., S. Behera, R. Arora, and S. Kumar (2017) Evolutionary adaptation of Kluyveromyces marxianus NIRE-K3 for enhanced xylose utilization. Front. Energy Res. 5: 32.
Pitkanen, J. P, A. Aristidou, L. Salusjarvi, L. Ruohonen, and M. Penttila (2003) Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab. Eng. 5: 16–31.
Fowler, Z. L., E. Leonard, and M. Koffas (2010) Biosynthesis of cellular building blocks: The prerequisites of life. pp. 3: 1–32. In: C. D. Smolke (ed.). The Metabolic Pathway Engineering Handbook. CRC Press, Boca Raton, FL, USA.
Fonseca, G. G., A. K. Gombert, E. Heinzle, and C. Wittmann (2007) Physiology of the yeast Kluyveromyces marxianus during batch and chemostat cultures with glucose as the sole carbon source. FEMS Yeast Res. 7: 422–435.
Stephanopoulos, G., A. A. Aristidou, and J. Nielsen (1998) Metabolic Engineering: Principles and Methodologies. Academic Press, San Diego, CA, USA.
Sauer, U., D. R. Lasko, J. Fiaux, M. Hochuli, R. Glaser, T. Szyperski, K. Wuthrich, and J. E. Bailey (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181: 6679–6688.
Arora, R., S. Behera, N. K. Sharma, and S. Kumar (2017) Augmentation of ethanol production through statistically designed growth and fermentation medium using novel thermotolerant yeast isolates. Renew Energy. 109: 406–421.
Behera, S., R. Arora, N. K. Sharma, and S. Kumar (2014) Fermentation of glucose and xylose sugar for the production of ethanol and xylitol by the newly isolated NIRE-GX1 yeast. pp. 175–182. In: S. Kumar, A. K. Sarma, S. K. Tyagi, and Y. K. Yadav (eds.). Recent Advances in Bio-energy Research Vol. III. SSS-NIRE, Kapurthala, India.
Tanimura, A., T. Nakamura, I. Watanabe, J. Ogawa, and J. Shima (2012) Isolation of a novel strain of Candida shehatae for ethanol production at elevated temperature. SpringerPlus. 1: 27.
Ozcan, S. and M. Johnston (1999) Function and regulation of yeast hexose transporters. Microbiol. Mol Biol Rev. 63: 554–569.
Sharma, N. K., S. Behera, R. Arora, S. Kumar, and R. K. Sani (2018) Xylose transport in yeast for lignocellulosic ethanol production: current status. J. Biosci. Bioeng. 125: 259–267.
Ozcan, S. and M. Johnston (1995) Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell Biol. 15: 1564–1572.
Ozcan, S. and M. Johnston (1996) Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell Biol. 16: 5536–5545.
Serrano, R. and G. Delafuente (1974) Regulatory properties of the constitutive hexose transport in Saccharomyces cerevisiae. Mol. Cell Biochem. 5: 161–171.
Zhang, Y., Z. Huang, C. Du, Y. Li, and Z. Cao (2009) Introduction of an NADH regeneration system into Klebsiella oxytoca leads to an enhanced oxidative and reductive metabolism of glycerol. Metab. Eng. 11: 101–106.
Hou, J., G. Scalcinati, M. Oldiges, and G. N. Vemuri (2010) Metabolic impact of increased NADH availability in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 76: 851–859.
Geertman, J. M., J. P. van Dijken, and J. T. Pronk (2006) Engineering NADH metabolism in Saccharomyces cerevisiae: formate as an electron donor for glycerol production by anaerobic, glucose-limited chemostat cultures. FEMS Yeast Res. 6: 1193–1203.
Nissen, T. L., C. W. Hamann, M. C. Kielland-Brandt, J. Nielsen, and J. Villadsen (2000) Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 16: 463–474.
Jain, V. K., B. Divol, B. A. Prior, and F. F. Bauer (2012) Effect of alternative NAD+-regenerating pathways on the formation of primary and secondary aroma compounds in a Saccharomyces cerevisiae glycerol-defective mutant. Appl. Microbiol. Biotechnol. 93: 131–141.
Mayer, G., K. D. Kulbe, and B. Nidetzky (2002) Utilization of xylitol dehydrogenase in a combined microbial/enzymatic process for production of xylitol from D-glucose. Appl. Biochem. Biotechnol. 98: 577–589.
Sakakibara, Y., B. C. Saha, and P. Taylor (2009) Microbial production of xylitol from L-arabinose by metabolically engineered Escherichia coli. J. Biosci. Bioeng. 107: 506–511.
Li, H., J. Su, W. Ma, A. Guo, Z. Shan, and H. Wang (2015) Metabolic flux analysis of Saccharomyces cerevisiae in a sealed winemaking fermentation system. FEMS Yeast Res. 15: fou010.
Hayakawa, K., F. Matsuda, and H. Shimizu (2018) 13C-metabolic flux analysis of ethanol-assimilating Saccharomyces cerevisiae for S-adenosyl-L-methionine production. Microb. Cell Fact. 17: 82.
Pfau, T., M. P. Pacheco, and T. Sauter (2016) Towards improved genome-scale metabolic network reconstructions: unification, transcript specificity and beyond. Brief Bioinform. 17: 1060–1069.
Ravikrishnan, A. and K. Raman (2015) Critical assessment of genome-scale metabolic networks: the need for a unified standard. Brief Bioinform. 16: 1057–1068.
Tomas-Gamisans, M., P. Ferrer, and J. Albiol (2016) Integration and validation of the genome-scale metabolic models of Pichia pastoris: A comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PLoS One. 11: e0148031.
Marcisauskas, S., B. Ji, and J. Nielsen (2019) Reconstruction and analysis of a Kluyveromyces marxianus genome-scale metabolic model. BioRxiv. 581587.
Ghosh, A., H. Zhao, and N. D. Price (2011) Genome-scale consequences of cofactor balancing in engineered pentose utilization pathways in Saccharomyces cerevisiae. PLoS One. 6: e27316.
Bordel, S., R. Agren, and J. Nielsen (2010) Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput. Biol. 6: e1000859.
Acknowledgements
The authors acknowledge the financial support provided by Ministry of New and Renewable Energy, Govt. of India to pursue the research activities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Arora, R., Behera, S., Sharma, N.K. et al. Evaluating the Pathway for Co-fermentation of Glucose and Xylose for Enhanced Bioethanol Production Using Flux Balance Analysis. Biotechnol Bioproc E 24, 924–933 (2019). https://doi.org/10.1007/s12257-019-0026-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-019-0026-5