Abstract
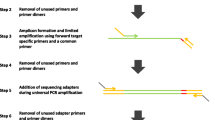
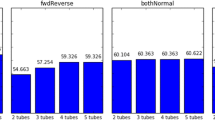
The multiplex PCR is one of the important methods to enrich the target DNAs for next generation sequencing. The non-specific amplification and interaction between the primers are the pivotal challenges of multiplex PCR. Here, we introduce the novel blunt hairpin primers for effective reducing the primer dimers and mispriming events. We also used a pair of auxiliary primers to enhance PCR efficiency. We simultaneously amplified 89 target regions from 44 samples and sequenced all amplicons on ion torrent PGM platform. Among all the filtrated amplicons (3438 different amplicons), 99.7, 97.6, 90.1 and 72.8% had sequencing depths fell within 200, 100, 50 and 25-fold range. The sequencing depth variations among all the samples were less than 27-fold. We also amplified multiplex regions with blunt hairpin, stick hairpin and normal linear primers, and the blunt hairpin primers could significantly reduce the amount of primer dimers and unspecific products.These results show that multiplex PCR with the blunt hairpin primers is a flexible, specific and economical target-region captured approach for the next generation sequencing.
Similar content being viewed by others
References
Mamanova, L., A. J. Coffey, C. E. Scott, I. Kozarewa, E. H. Turner, A. Kumar, E. Howard, J. Shendure, and D. J. Turner (2010) Target-enrichment strategies for next-generation sequencing. Nat. Meth. 7: 111–118.
Varley, K. E. and R. D. Mitra (2008) Nested Patch PCR enables highly multiplexed mutation discovery in candidate genes. Genome Res. 18: 1844–1850.
Dahl, F., M. Gullberg, J. Stenberg, U. Landegren, and M. Nilsson (2005) Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 33: e71.
Dahl, F., J. Stenberg, S. Fredriksson, K. Welch, M. Zhang, M. Nilsson, D. Bicknell, W. F. Bodmer, R. W. Davis, and H. Ji (2007) Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Nat. Acad. Sci. USA. 104: 9387–9392.
Meuzelaar, L. S., O. Lancaster, J. P. Pasche, G. Kopal, and A. J. Brookes (2007) MegaPlex PCR: a strategy for multiplex amplification. Nat. Meth. 4: 835–837.
Teer, J. K., L. L. Bonnycastle, P. S. Chines, N. F. Hansen, N. Aoyama, A. J. Swift, H. O. Abaan, T. J. Albert, N. C. S. Program, E. H. Margulies, E. D. Green, F. S. Collins, J. C. Mullikin, and L. G. Biesecker (2010) Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Gen. Res. 20: 1420–1431.
Kaboev, O. K., L. A. Luchkina, A. N. Tret'iakov, and A. R. Bahrmand (2000) PCR hot start using primers with the structure of molecular beacons (hairpin-like structure). Nucleic Acids Res. 28: E94.
Ailenberg, M. and M. Silverman (2000) Controlled hot start and improved specificity in carrying out PCR utilizing touch-up and loop incorporated primers (TULIPS). BioTechniques 29: 1018–1020, 1022-1024.
Nazarenko, I., B. Lowe, M. Darfler, P. Ikonomi, D. Schuster, and A. Rashtchian (2002) Multiplex quantitative PCR using selfquenched primers labeled with a single fluorophore. Nucleic Acids Res. 30: e37.
Stahlberg, A., P. M. Krzyzanowski, J. B. Jackson, M. Egyud, L. Stein, and T. E. Godfrey (2016) Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 44: e105.
Nguyen-Dumont, T., B. J. Pope, F. Hammet, M. C. Southey, and D. J. Park (2013) A high-plex PCR approach for massively parallel sequencing. BioTechniques. 55: 69–74.
Han, J. (2013) Multiplex PCr in molecular differential diagnosis of microbial infections: methods, utility, and platforms. pp. 627–646. Advanced Techniques in Diagnostic Microbiology. Springer, City.
Nguyen-Dumont, T., F. Hammet, M. Mahmoodi, B. J. Pope, G. G. Giles, J. L. Hopper, M. C. Southey, and D. J. Park (2015) Abridged adapter primers increase the target scope of Hi-Plex. BioTechniques. 58: 33–36.
Chen, K., Y. X. Zhou, K. Li, L. X. Qi, Q. F. Zhang, M. C. Wang, and J. H. Xiao (2016) A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal. Bioanal. Chem. 408: 4371–4377.
Depristo, M. A. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Gen. 43: 491–498.
Mokry, M., H. Feitsma, I. J. Nijman, E. de Bruijn, P. J. van der Zaag, V. Guryev, and E. Cuppen (2010) Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Res. 38: e116.
Nielsen, R., J. S. Paul, A. Albrechtsen, and Y. S. Song (2011) Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Gen. 12: 443–451.
Campbell, N. R., S. A. Harmon, and S. R. Narum (2015) Genotyping-in-Thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Res. 15: 855–867.
Pan, W., M. Byrne-Steele, C. Wang, S. Lu, S. Clemmons, R. J. Zahorchak, and J. Han (2014) DNA polymerase preference determines PCR priming efficiency. BMC Biotechnol. 14: 10.
Craig, D. W., J. V. Pearson, S. Szelinger, A. Sekar, M. Redman, J. J. Corneveaux, T. L. Pawlowski, T. Laub, G. Nunn, D. A. Stephan, N. Homer, and M. J. Huentelman (2008) Identification of genetic variants using bar-coded multiplexed sequencing. Nature Meth. 5: 887–893.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Chen, K., Zhou, Y., Li, K. et al. Multiplex PCR with the blunt hairpin primers for next generation sequencing. Biotechnol Bioproc E 22, 347–351 (2017). https://doi.org/10.1007/s12257-017-0133-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-017-0133-0