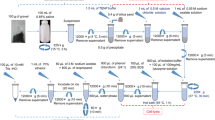
Abstract
We demonstrated ultrasonication as a rapid and high yield DNA extraction method suitable for bacterial gene quantification by NanoGene assay. The NanoGene assay utilizes DNA hybridization in solution and a combination of magnetic beads and quantum dot nanoparticles. Unlike the existing gene quantification assays, the NanoGene method is capable of quantifying genes in the presence of environmental inhibitors and cell materials. The performance of the ultrasonication was compared with heating and freeze-thaw. They first were evaluated for their cell lysis capability in humic acids laden sand samples, via EtBr assay. Using autoclaved samples as a bench mark, their cell lysis capability were 106 ± 3, 68 ± 5, and 48 ± 15%, respectively. Morphological changes of cells for each method were also observed by FE-SEM. More importantly, ultrasonication performed significantly better (more than 3× fluorescence signal) than commercial DNA extraction methods during bacterial gene quantification in humic acids laden sand samples.
Similar content being viewed by others
References
Liu, G., F. Q. Ling, A. Magic-Knezev, W. T. Liu, J. Q. J. C. Verberk, and J. C. Van Dijk (2013) Quantification and identification of particle-associated bacteria in unchlorinated drinking water from three treatment plants by cultivation-independent methods. Water Res. 47: 3523–3533.
Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler (2002) Real-time PCR quantification of nitrifying nacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37: 343–351.
El Fantroussi, S. and S. N. Agathos (2005) Is bioaugmentation a feasible strategy for pollutant removal and site remediation? Curr. Opin. Microbiol. 8: 268–275.
Kao, C. -M., C. S. Chen, F. -Y. Tsa, K. -H. Yang, C. -C. Chien, S. -H. Liang, C. -A. Yang, and S. C. Chen (2010) Application of realtime PCR, DGGE fingerprinting, and culture-based method to evaluate the effectiveness of intrinsic bioremediation on the control of petroleum-hydrocarbon plume. J. Hazard. Mater. 178: 409–416.
Chon, K., J. -S. Chang, E. Lee, J. Lee, J. Ryu, and J. Cho (2011) Abundance of denitrifying genes coding for nitrate (narG), nitrite (nirS), and nitrous oxide (nosZ) reductases in estuarine versus wastewater effluent-fed constructed wetlands. Ecol. Eng. 37: 64–69.
Sims, A., J. Horton, S. Gajaraj, S. McIntosh, R. J. Miles, R. Mueller, R. Reed, and Z. Hu (2012) Temporal and spatial distributions of ammonia-oxidizing archaea and bacteria and their ratio as an indicator of oligotrophic conditions in natural wetlands. Water Res. 46: 4121–4129.
Looper, J. K., A. Cotto, B. -Y. Kim, M. -K. Lee, M. R. Liles, S. M. N. Chadhainf, and A. Son (2013) Microbial community analysis of Deepwater Horizon oil-spill impacted sites along the Gulf coast using functional and phylogenetic markers. Environ. Sci. Proc. Impact. 15: 2068–2079.
Kim, G. -Y. and A. Son (2010) Development and characterization of a magnetic bead-quantum dot nanoparticles based assay capable of Escherichia coli O157:H7 quantification. Anal. Chim. Act. 677: 90–96.
Lee, S. Y., J. Bollinger, D. Bezdicek, and A. Ogram (1996) Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62: 3787–3793.
Mumy, K. L. and R. H. Findlay (2004) Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. J. Microbiol. Meth. 57: 259–268.
Peršoh, D., S. Theuerl, F. Buscot, and G. Rambold (2008) Towards a universally adaptable method for quantitative extraction of high-purity nucleic acids from soil. J. Microbiol. Meth. 75: 19–24.
Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson (1998) Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64: 2463–2472.
Lakay, F. M., A. Botha, and B. A. Prior (2007) Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. J. Appl. Microbiol. 102: 265–273.
Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse (1999) Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65: 4715–4724.
Howeler, M., W. C. Ghiorse, and L. P. Walker (2003) A quantitative analysis of DNA extraction and purification from compost. J. Microbiol. Meth. 54: 37–45.
Robe, P., R. Nalin, C. Capellano, T. M. Vogel, and P. Simonet (2003) Extraction of DNA from soil. Eur. J. Soil. Sci. 39: 183–190.
Zhou, J., M. A. Bruns, and J. M. Tiedje (1996) DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62: 316–322.
Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen (1994) Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60: 1572–1580.
Parachin, N., J. Schelin, B. Norling, P. Rådström, and M. Gorwa-Grauslund (2010) Flotation as a tool for indirect DNA extraction from soil. Appl. Microbiol. Biotechnol. 87: 1927–1933.
Steffan, R. J., J. Goksoyr, A. K. Bej, and R. M. Atlas (1988) Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54: 2908–2915.
Tsai, Y. L. and B. H. Olson (1992) Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl. Environ. Microbiol. 58: 754–757.
Mitchell, K., B. Chua, and A. Son (2014) Development of first generation in-situ pathogen detection system (Gen1-IPDS) based on NanoGene assay for near real time E. coli O157:H7 detection. Biosens. Bioelectronics. 54: 229–236.
Cébron, A., M.-P. Norini, T. Beguiristain, and C. Leyval (2008) Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods. 73: 148–159.
Wang, X., B. -T. Lee, and A. Son (2014) Physical lysis only (PLO) methods suitable as rapid sample pretreatment for qPCR assay. Appl. Microbiol. Biotechnol. 98: 8719–8728.
Rodrigues, L., J. Ramos, I. Couto, L. Amaral, and M. Viveiros (2011) Ethidium bromide transport across Mycobacterium smegmatis cell-wall: Correlation with antibiotic resistance. BMC Microbiol. 11.
Dragan, A. I., E. S. Bishop, R. J. Strouse, J. R. Casas-Finet, M. A. Schenerman, and C. D. Geddes (2009) Metal-enhanced ethidium bromide emission: Application to dsDNA detection. Chem. Phys. Lett. 480: 296–299.
Olmsted, J. I. and D. R. Kearns (1977) Mechanism of ethidium bromide fluorescence enhancement on bing to nucleic acids. Biochem. 16: 3647–3654.
Waring, M. J. (1965) Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 13: 269–282.
Brum, M. C. and J. F. Oliveira (2007) Removal of humic acid from water by precipitate flotation using cationic surfactants. Miner Eng. 20: 945–949.
Bapat, B. S., P. R. Gogate, and A. B. Pandit (2008) Theoretical analysis of sonochemical degradation of phenol and its chloroderivatives. Ultrason. Sonochem. 15: 564–570.
Chowdhury, P. and T. Viraraghavan (2009) Sonochemical degradation of chlorinated organic compounds, phenolic compounds and organic dyes -a review. Sci. Total. Environ. 407: 2472–2492.
Im, J. -K., L. K. Boateng, J. R. V. Flora, N. Her, K. -D. Zoh, A. Son, and Y. Yoon (2014) Enhanced ultrasonic degradation of acetaminophen and naproxen in the presence of powered activated carbon and biochar adsorbents. Sep. Purif. Technol. 123: 96–106.
Wang, Y. F., D. Zhao, W. H. Ma, C. C. Chen, and J. C. Zhao (2008) Enhanced sonocatalytic degradation of azo dyes by Au/TiO2. Environ. Sci. Technol. 42: 6173–6178.
Loza, V., E. Perona, and P. Mateo (2013) Molecular fingerprinting of cyanobacteria from river biofilms as a water quality monitoring tool. Appl. Environ. Microbiol. 79: 1459–1472.
de Lipthay, J. R., C. Enzinger, K. Johnsen, J. Aamand, and S. J. Sørensen (2004) Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol. Biochem. 36: 1607–1614.
Krsek, M. and E. M. H. Wellington (1999) Comparison of different methods for the isolation and purification of total community DNA from soil. J. Microbiol. Meth. 39: 1–16.
Frostegård, Å., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet (1999) Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65: 5409–5420.
Westergaard, K., A. K. Müller, S. Christensen, J. Bloem, and S. J. Sørensen (2001) Effects of tylosin as a disturbance on the soil microbial community. Soil Biol. Biochem. 33: 2061–2071.
Esteban, J., N. Alonso-Rodriguez, G. del-Prado, A. Ortiz-Pérez, D. Molina-Manso, J. Cordero-Ampuero, E. Sandoval, R. Fernández-Roblas, and E. Gómez-Barrena (2012) PCR-hybridization after sonication improves diagnosis of implant-related infection. Acta Orthop. 83: 299–304.
Bollet, C., M. J. Gevaudan, X. de Lamballerie, C. Zandotti, and P. de Micco (1991) A simple method for the isolation of chromosomal DNA from Gram positive or acid-fast bacteria. Nucleic Acids Res. 19: 1955.
Ahmed, O. B., A. H. Asghar, and M. M. Elhassan (2014) Comparison of three DNA extraction methods for polymearase chain reaction (PCR) analysis of bacterial genomic DNA. Afr. J. Microbiol. Res. 8: 598–602.
Tsai, Y. L. and B. H. Olson (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57: 1070–1074.
Chen, N. T. and C. W. Chang (2012) Quantification of Legionella pneumophila by real-time quantitative PCR from samples with humic acid and ferric ion. Sci. Total Environ. 414: 608–613.
Wang, X. and A. Son (2013) Effects of pretreatment on the denaturation and fragmentation of genomic DNA for DNA hybridization. Environ. Sci. Proc. Impact. 15: 2204–2212.
Brautigam, A. R., D. D. Richman, and M. N. Oxman (1980) Rapid typing of herpes simplex virus isolates by deoxyribonucleic acid: Deoxyribonucleic acid hybridization. J. Clin. Microbiol. 12: 226–234.
Tebbe, C. C. and W. Vahjen (1993) Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59: 2657–2665.
Wilson, I. (1997) Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63: 3741–3751.
Wang, X., M. R. Liles, and A. Son (2013) Quantification of E. coli O157:H7 in soils using an inhibitor-resistant NanoGene assay. Soil Biol. Biochem. 58: 9–15.
Kim, G. -Y., X. Wang, and A. Son (2011) Inhibitor resistance and in situ capability of nanoparticle based gene quantification. J. Environ. Monitor. 13: 1344–1350.
Kim, G. -Y., X. Wang, H. Ahn, and A. Son (2011) Gene quantification by the NanoGene assay is resistant to inhibition by humic acids. Environ. Sci. Technol. 45: 8873–8880.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, X., Cho, KS. & Son, A. Ultrasonication as a rapid and high yield DNA extraction method for bacterial gene quantification by NanoGene assay. Biotechnol Bioproc E 20, 1133–1140 (2015). https://doi.org/10.1007/s12257-015-0465-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-015-0465-6