Abstract
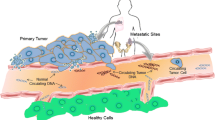
The use of circulating tumor DNA (ctDNA) as a prognostic and/or predictive biomarker has been proven in numerous studies. Recent technical advancements have improved sensitivity, specificity, and feasibility of ctDNA detection enabling innovative clinical applications. Besides its potential use as a diagnostic biomarker and the detection of recurrence or minimal residual disease, the most widespread application of the so-called liquid biopsy is real-time monitoring of treatment response and tumor evolution. Since tissue biopsies provide just a static snapshot of the tumor at the time of biopsy and do not necessarily represent the entire tumor genome, a sequential analysis of ctDNA enables an early identification of resistance mechanisms before they become clinically obvious. Furthermore, novel therapy targets that have not been present in available tumor samples might be identified. However, for an actual implementation of the liquid biopsy in clinical practice, it is essential to develop standardized pre-analytical and analytical methodologies and to resolve some outstanding question with respect to origin, biology, and dynamics of ctDNA. In this short review, the clinical utility and existing limitations of ctDNA focusing on monitoring treatment response will be discussed.


Similar content being viewed by others
References
Schilsky RL. Implementing personalized cancer care. Nat Rev Clin Oncol. 2014;11(7):432–8. PubMed PMID: 24687035.
Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational Implications of tumor heterogeneity. Clin Cancer Res. 2015;21(6):1258–66. PubMed PMID: 25770293. Pubmed Central PMCID: 4374162.
Gerlinger M. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Mandel PMP. Les acides nucleiques du plasma sanguin chez l’homme [in French]. Seances Soc Biol Fil. 1948;142:241–3.
Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–50. PubMed PMID: 837366.
Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23. PubMed PMID: 25388429.
Cirkel GA, Gadellaa-van Hooijdonk CG, Koudijs MJ, Willems SM, Voest EE. Tumor heterogeneity and personalized cancer medicine: are we being outnumbered? Future Oncol. 2014;10(3):417–28. PubMed PMID: 24559448.
Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86. PubMed PMID: 24449238. Epub 2014/01/23. Eng.
Gonzalez-Masia JA, Garcia-Olmo D, Garcia-Olmo DC. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 2013;6:819–32. PubMed PMID: 23874104. Pubmed Central PMCID: 3711950.
Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014;86(3):136–42. PubMed PMID: 24761422. Epub 2014/04/25. Eng.
Madhavan D, Wallwiener M, Bents K, Zucknick M, Nees J, Schott S, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146(1):163–74. PubMed PMID: 24838941.
Chen X. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin Cancer Res. 1999;5:2297–303.
Sozzi G. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61:4675–8.
Arnalich F, Menendez M, Lagos V, Ciria E, Quesada A, Codoceo R, et al. Prognostic value of cell-free plasma DNA in patients with cardiac arrest outside the hospital: an observational cohort study. Crit Care. 2010;14(2):R47. PubMed PMID: 20350299. Pubmed Central PMCID: 2887159.
Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10(2):R60. PubMed PMID: 16613611. Pubmed Central PMCID: 1550922.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. PubMed PMID: 24553385. Epub 2014/02/21. Eng.
Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res. 2014;16(4):421. PubMed PMID: 25107527.
Heitzer E, Auer M, Hoffmann EM, Pichler M, Gasch C, Ulz P, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer. 2013;133(2):346–56. PubMed PMID: 23319339. Pubmed Central PMCID: 3708119.
Thierry AR, Mouliere F, Messaoudi SE, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–5. PubMed PMID: 24658074. Epub 2014/03/25. Eng.
Misale S. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6.
Kuo YB, Chen JS, Fan CW, Li YS, Chan EC. Comparison of KRAS mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clin Chim Acta. 2014;433:284–9. PubMed PMID: 24685572.
Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, Auer M, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014;10(3):e1004271. PubMed PMID: 24676216. Pubmed Central PMCID: 3967949.
Diaz LA Jr. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40.
Thijssen MA, Swinkels DW, Ruers TJ, de Kok JB. Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res. 2002;22:421–5.
Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731–6. PubMed PMID: 25628445. Pubmed Central PMCID: 4374387.
Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5(4):30. PubMed PMID: 23561577. Pubmed Central PMCID: 3707016.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. PubMed PMID: ISI:000258988600033. English.
Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–209. PubMed PMID: 23484797.
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12. PubMed PMID: 23563269.
Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. PubMed PMID: 23197571. Pubmed Central PMCID: 3641759.
Reinert T, Scholer LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2015. PubMed PMID: 25654990.
Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–65. PubMed PMID: 25185240.
Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-SEq. Expert Rev Mol Diagn. 2015;16:1–5. PubMed PMID: 25773944.
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54. PubMed PMID: 24705333. Epub 2014/04/08. Eng.
Kidess E, Heirich K, Wiggin M, Vysotskaia V, Visser BC, Marziali A, et al. Mutation profiling of tumor DNA from plasma and tumor tissue of colorectal cancer patients with a novel, high-sensitivity multiplexed mutation detection platform. Oncotarget. 2015;6(4):2549–61. PubMed PMID: 25575824.
Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA. 2015;112(11):E1317–25. PubMed PMID: 25646427. Pubmed Central PMCID: 4372002.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heitzer, E. Clinical utility of circulating tumor DNA in human cancers. memo 8, 222–226 (2015). https://doi.org/10.1007/s12254-015-0217-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12254-015-0217-5