Abstract
The major aim of this study was to evaluate the performance of anti-BRAF V600E (VE1) antibody in colorectal tumors with and without KRAS mutation. KRAS and BRAF are two major oncogenic drivers of colorectal cancer (CRC) that have been frequently described as mutually exclusive, thus the BRAF V600E mutation is not expected to be present in the cases with KRAS mutation. In addition, a review of 25 studies comparing immunohistochemistry (IHC) using the anti-BRAF V600E (VE1) antibody with BRAF V600E molecular testing in 4041 patient samples was included.
One-hundred and twenty cases with/without KRAS or BRAF mutations were acquired. The tissue were immunostained with anti-BRAF V600E (VE1) antibody with OptiView DAB IHC detection kit. The KRAS mutated cases with equivocal immunostaining were further evaluated by Sanger sequencing for BRAF V600E mutation. Thirty cases with BRAF V600E mutation showed unequivocal, diffuse, uniform, positive cytoplasmic staining and 30 cases with wild-type KRAS and BRAF showed negative staining with anti-BRAF V600E (VE1) antibody. Out of 60 cases with KRAS mutation, 56 cases (93.3%) were negative for BRAF V600E mutation by IHC. Four cases showed weak, equivocal, heterogeneous, cytoplasmic staining along with nuclear staining in 25–90% of tumor cells. These cases were confirmed to be negative for BRAF V600E mutation by Sanger sequencing. Overall, IHC with anti-BRAF V600E (VE1) antibody using recommended protocol with OptiView detection is optimal for detection of BRAF V600E mutation in CRC. Our data are consistent with previous reports indicating that KRAS and BRAF V600E mutation are mutually exclusive.
Similar content being viewed by others
Introduction
Colorectal cancer is the third most common cancer and the fourth most prevalent cause of death in the world [46]. Approximately 35–45% of patients with colorectal tumors have mutation in KRAS gene, while BRAF V600E mutation is found in about 5–15% of colorectal adenocarcinomas [8, 9, 26, 31]. Both these mutations are considered to be oncogenic driver mutations, since they are both responsible for the initiation and maintenance of the tumor [10]. Importantly, many studies have indicated that BRAF V600E mutation occurs only in tumors that do not carry mutations in KRAS gene and it is widely accepted that these two mutations are mutually exclusive [10, 23, 25, 30].
The BRAF gene encodes a cytoplasmic serine-threonine kinase that is frequently mutated in various cancers, including melanoma, papillary thyroid carcinoma, and colorectal carcinoma, among others. The oncogenic mutations in BRAF gene result in constitutive activation of the MAPK signaling pathway, leading to increased cell proliferation, resistance to apoptosis and tumor progression. The most common of these mutations, the V600E mutation, occurs in exon 15 and results in a substitution from valine to glutamic acid at position 600 within the BRAF kinase domain.
BRAF V600E mutation occurs in about 5% of microsatellite stable (MSS) CRC tumors. These tumors are associated with a distinct molecular and clinical phenotype with a poor prognosis [40]. The presence of BRAF V600E mutation in CRC is associated with poor survival [44]. BRAF V600E mutation is also detected in sporadic CRC tumors with microsatellite instability (MSI) [27]. Particularly, BRAF V600E mutation is observed in about two thirds of MSI tumors with the loss of MLH1 expression due to MLH1 promoter methylation [18]. In contrast, BRAF V600E mutation is very rare in CRC patients with Lynch syndrome [27]. In clinical practice it is much easier to detect BRAF V600E mutation than methylation status of MLH1 promoter [26]. Therefore, it was suggested that assessment of BRAF V600E mutation can be used to triage patients for mismatch repair (MMR) genetic testing to differentiate MLH1-deficient sporadic CRC from Lynch syndrome caused by germ-line MLH1 mutations [7, 14, 17, 26, 41, 43]. Currently, the American National Comprehensive Cancer Network (NCCN) guidelines recommend that BRAF V600E mutational status should be evaluated in all colorectal carcinomas to identify 1) the patients with Lynch syndrome in MMR deficient group and 2) to identify the MMR proficient/BRAF V600E group with poor prognosis [15, 38, 43].
The most common approach for the detection of BRAF mutation is sequencing of tumor DNA, for example Sanger sequencing, pyrosequencing and high resolution melting. All of these methods are able to detect a mutant allele in a background of 5–20 fold excess of wild-type alleles. In contrast, immunohistochemistry allows direct visualization of the mutant protein in the tumor cells in tissue context. The anti-BRAF V600E (VE1) antibody is currently used to evaluate the BRAF V600E mutation status in various cancers including CRC [32]. This antibody is a mutation-specific mouse monoclonal antibody that was raised against a synthetic peptide representing the BRAF V600E mutated amino acid sequence from amino acids 596 to 606 (GLATEKSRWSG) [5, 6].
The primary goal of this study was to compare the performance of the anti-BRAF V600E (VE1) antibody to detect BRAF V600E mutation by IHC in colon cancer cases with/without KRAS mutation. Since concomitant KRAS and BRAF tumor mutations are considered mutually exclusive we wanted to confirm that the CRC cases carrying KRAS mutation show negative BRAF V600E staining by IHC with anti-BRAF V600E (VE1) antibody. In addition, we performed a review of 25 studies that compared IHC using anti-BRAF V600E (VE1) antibody with molecular testing for BRAF V600E mutation.
Materials and Methods
Tumor Specimens
A total of 120 formalin-fixed paraffin embedded (FFPE) tissues from patients with colorectal cancer were ordered from Avaden Biosciences and GLAS/Consultants in Human Biologics. The requested cases included 60 CRC cases with confirmed KRAS mutation, 30 CRC cases with confirmed BRAF V600E mutation and 30 CRC cases confirmed to be wild-type BRAF and wild-type KRAS. The presence/absence of these mutations was confirmed by molecular testing by the vendor.
BRAF V600E Immunohistochemistry
Four 4 μm thick sections were cut from the FFPE blocks. The testing was performed using anti-BRAF V600E (VE1) mouse monoclonal primary antibody (Ventana Medical Systems, Inc., Cat. Number 790–4855) the BenchMark ULTRA platform with Cell Conditioning 1 for 64 min, pre-peroxidase inhibition and primary antibody incubation for 16 min at 37 °C. Final concentration of the antibody was ~12 μg/ml. The OptiView DAB IHC Detection Kit (Ventana Medical Systems, Inc.) was used to detect BRAF V600E protein expression. Tissues were counterstained with Hematoxylin II (Ventana Medical Systems, Inc.) and Bluing Reagent (Ventana Medical Systems, Inc.) for 4 min. To measure the level of non-specific background signal, each tissue was also stained with a mouse monoclonal antibody (MOPC-21) [Negative Control (Monoclonal), Ventana Medical Systems, Inc.]. This antibody is not directed against any known epitope present in human tissue. In addition, slides containing 2 cases positive for BRAF V600E mutation (CRC, thyroid papillary carcinoma) and one case negative for BRAF V600E mutation (CRC) were used as run control slides. The absence/presence of the BRAF V600E mutation in the tissues was confirmed by Sanger sequencing. These slides were included with each individual run to assess the expected quality of the antibody and all components of the assay. The overall run was accepted if: 1) the BRAF V600E positive tissue control stained with anti-BRAF V600E (VE1) antibody showed specific cytoplasmic staining pattern and had acceptable background; 2) the positive tissue control stained with Negative Control Monoclonal shows no specific staining and had acceptable background; 3) the BRAF V600E negative tissue control stained with anti-BRAF V600E (VE1) antibody showed no specific staining pattern and acceptable background; and 4) the BRAF V600E negative tissue control stained with Negative Control (Monoclonal) shows no specific staining and has acceptable background.
The stain intensity of anti-BRAF V600E (VE1) antibody in tumor cells was recorded on a 0–3 scale. Strong cytoplasmic staining was scored as 3, medium cytoplasmic staining as 2, weak cytoplasmic staining as 1 and the absence of staining was scored as 0. In addition, any nuclear staining and the percentage of tumor cells stained positive with anti-BRAF V600E (VE1) antibody was recorded. The criteria for positive BRAF V600E staining included unequivocal, diffuse, uniform, cytoplasmic staining at intensity ≥1 in majority of malignant cells. The cases were scored as negative for BRAF V600E mutation if they showed no staining or weak, cytoplasmic, non-granular, uniform staining (stain intensity <1). The cases with staining of isolated tumor cells in a tumor that otherwise showed no staining were also scored as negative. The cases were scored as equivocal if they displayed ambiguous, heterogeneous, non-uniform cytoplasmic staining in tumor cells with or without nuclear staining. The equivocal cases were sequenced by Sanger sequencing to confirm the presence/absence of BRAF V600E mutation.
BRAF V600E Sanger Sequencing
Genomic DNA was extracted from 20 μm thick sections from FFPE samples using the QIAamp FFPE Tissue Kit (Qiagen, Redwood, CA) according to manufacturer’s instructions. The primers for Sanger sequencing were designed to amplify region of the exon 15 of the BRAF gene coding sequences at mutation site and a few nucleotides in the intron on both ends. Two primers were used including 1) BRAF-ex15F-TGCTTGCTCTGATAGGAAAATG and 2) BRAF-ex15R-AGCATCTCAGGGCCAAAAAT. Both forward and reverse strands were sequenced on an Applied Biosystem’s 3730xl DNA Analyzer and analyzed using DNASTAR Lasergene 12 software (DNASTAR, Madison, WI).
Results
Anti-BRAF V600E (VE1) Immunohistochemistry
All 120 cases were examined for presence of BRAF V600E mutation by IHC using the anti-BRAF V600E (VE1) antibody on the automated VENTANA BenchMark ULTRA platform.
All 30 cases with BRAF V600E mutation exhibited uniform, unequivocal, diffuse, cytoplasmic staining in majority of tumor cells with stain intensities of 1–2.75 and background ≤0.25. Out of 30 cases, 28 cases showed positive BRAF V600E signal in 100% of tumor cells and 2 cases showed positive staining in 90% and 85% of tumor cells, respectively. These data are consistent with previous reports indicating that majority of tumor cells express mutated BRAF V600E protein, since this mutation is driving tumor proliferation. All 30 cases with no BRAF V600E and no KRAS mutations showed intensities of ≤0.5 and background ≤0.25. In 28 cases the stain intensities were 0–0.25, in remaining two cases 10% and 85% of malignant cells stained at the stain intensities 0.5. The sensitivity and specificity was 100% for cases with confirmed BRAF V600E mutational status. Representative images are shown in Fig. 1.
Representative images of eight colon cancer cases stained with anti-BRAF V600E (VE1) mouse monoclonal antibody. BRAF V600E mutation was confirmed in cases shown in images A,B,E,F,I,J,M,N by molecular testing, no BRAF V600E mutation was detected by molecular testing in cases shown in images C,D,G,H,K,L,O,P. Magnification 10× (A,C,D,E,G,I,K,M,O) and 20× (B,D,F,H,J,L,N,P)
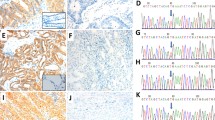
Out of 60 CRC cases with KRAS mutations 56 cases were scored as negative for BRAF V600E mutation (stain intensity <1). There were four cases where the stain intensities were scored as 1. However, these four cases exhibited ambiguous, heterogeneous, non-uniform cytoplasmic staining along with nuclear staining and thus they were scored as equivocal. In the first case, only 40–50% of tumor cells were positively stained with anti-BRAF V600E (VE1) antibody, the cells with positive staining showed the signal in cytoplasm and also strong signal in nuclei, the cytoplasmic staining was non-diffuse and non-uniform. Representative images are shown in Fig. 2A-C. In the second case only small portion of tumor showed uneven, cytoplasmic staining (25% tumor cells). In addition, tumor cells also exhibited nuclear staining. Representative images are shown in Fig. 2D-F. Third case showed staining in 70% of cells, however the staining was heterogeneous and clearly nuclear along with lighter non-uniform cytoplasmic staining. Representative images are shown in Fig. 2 G-I. Fourth case showed high degree of nuclear staining, overall 90% of tumor cells showed positive staining in cytoplasm, however the strong staining was observed in nuclei with some signal in cytoplasm. This cytoplasmic staining was scattered and uneven. Representative images are shown in Fig. 2J-L. Since these four case exhibited ambiguous, non-uniform, heterogeneous and nuclear staining pattern, they were assigned as equivocal for BRAF V600E mutation.
Representative images of four colon cancer cases with KRAS mutation showing equivocal staining. The tissues were stained with anti-BRAF V600E (VE1) mouse monoclonal antibody (A,B,D,E,G,H,J,K) and negative reagent control (C,F,I,L) [A,B,C - case 1; D,E,F - case 2; G,H,I - case 3; J,K,L - case 4, Magnification - 10× (A,D,G,J), 20× (B,C,E,F,H,I,K,L)]
In addition, there were 4 cases that were scored with stain intensity of 0.75 in all three evaluated slides in 30–90% of tumor cells. All these cases exhibited nuclear staining and non-diffuse, weak, heterogeneous cytoplasmic staining. Since the stain intensity was <1 these cases were scored as negative for BRAF V600E mutation.
DNA Sequencing
Overall, out of the all 120 cases, there were 60 cases with KRAS mutation. Out of these 60 cases, 4 cases showed BRAF V600E stain intensity 1 in 25–90% of tumor cells. This was not expected since these KRAS and BRAF V600E are mutually exclusive mutations. These cases exhibited the staining pattern that was not consistent with the recommendations based from studies shown in Table 1 that include uniform, diffuse, cytoplasmic staining in majority of malignant cells. In addition, 4 KRAS mutated cases showed stain intensities 0.75 with similar nuclear/heterogeneous staining pattern. These cases were also sequenced for BRAF V600E mutation. All these cases were negative for BRAF V600E mutation by Sanger sequencing.
Disscussion
This study evaluated 120 CRC cases with and without KRAS mutation to access the performance of IHC using anti-BRAF V600E (VE1) antibody for detection of BRAF V600E mutation. Overall, the results of these experiments demonstrates that IHC using the anti-BRAF V600E (VE1) antibody with the VENTANA OptiView DAB detection system and BenchMark ULTRA platform is a highly specific and sensitive method for the detection of BRAF V600E in colon cancer.
There is strong evidence from multiple studies that the IHC using anti-BRAF V600E (VE1) antibody is highly concordant with molecular tests for the BRAF V600E mutation. Table 2 shows a summary of 25 studies that evaluated sensitivity and specificity of IHC with anti-BRAF V600E (VE1) antibody in comparison with molecular tests using different methods (Sanger, pyroseqencing, SNapShot PCR, NGS, etc.). Altogether, 4041 patient samples were evaluated in these studies, the overall sensitivity and specificity of IHC assay using anti-BRAF V600E (VE1) antibody compared to molecular tests was 93% (934/1008) and 96% (2922/3033), respectively.
Out of these 25 studies, 4 publications reported lower sensitivity and/or specificity of anti-BRAF V600E antibody compared to sequencing [1, 12, 20, 21]. However, there were several problems with these studies. First, the study by Adackapara et al. analyzed 52 colorectal carcinomas with known BRAF mutation status determined by pyrosequencing and found that IHC had a low sensitivity (71%) and specificity (74%) for detecting BRAF V600E mutation compared to pyrosequencing (Table 2). They concluded that IHC using anti-BRAF V600E (VE1) antibody is not a useful surrogate for detecting BRAF mutation in colorectal carcinoma. However, in their experiment, manual staining with citrate buffer as antigen retrieval was employed. In our experience and the experience of others the use of acid for antigen retrieval step results in suboptimal staining that is difficult to interpret [19]. TRIS or EDTA buffers at pH = 8 proved to be retrieval agents that produced the most robust and homogenous cytoplasmic staining with anti-BRAF V600E (VE1) antibody. Similarly, Lasota et al. used in their studies Bond Epitope Retrieval Solution 1 (pH = 6) which is not an optimal solution for antigen retrieval for this assay [20].
Another important factor that may contribute to the different outcome of the studies is the interpretation of the IHC results. As multiple studies have highlighted, a proper scoring system is necessary to reduce false-positive and false-negative cases. Since BRAF V600E mutation is a driver mutation, a majority of tumor cells should express this mutated protein. The scoring criteria shown in Table 1 were used in the individual studies presented in Table 2 that compare IHC using anti-BRAF V600E (VE1) antibody with molecular testing for BRAF V600 E mutation. Overall, 14 out of 25 studies scored cases positive for BRAF V600E mutation when uniform or nearly uniform, diffuse staining was present in tumor cells or when the majority (≥ 75%) of tumor cells exhibited unequivocal cytoplasmic staining (Table 2). All studies that used these interpretation criteria (and appropriate protocol using antigen retrieval at alkaline pH) reached close to 100% sensitivity and specificity compared to sequencing. Additional 7 studies did not include scoring criteria in the method section, however they reported that homogeneous, diffuse staining pattern was observed in cases with confirmed BRAF V600E mutation. Three studies provided no details. In one study the cases were scored as positive for BRAF V600E staining when only ≥20% tumor cells showed positive signal in one study [12]. The sensitivity and specificity reported in this study was only 89% and 57% (when IHC assay on BenchMark ULTRA platform was used) and 75% and 93% (when IHC assay on Leica Bond was used). Several studies suggested that additional analysis is required for minority of equivocal cases with ambiguous, focal, heterogeneous staining (Table 2). False positive staining was noted in signet ring tumor cells [33]. Importantly, the nuclear staining was described as the most common artifact (Table 2) [22]. For example, Bledsoe et al. noted that BRAF-mutant cases showed homogeneous, finely granular, cytoplasmic staining with varying intensities, however, non-specific nuclear and heterogeneous non-diffuse cytoplasmic staining of variable intensity was observed in a minority of non-BRAF mutant cases [3]. Therefore, it was recommended by Marin et al. that “the interpretation should be made with caution in the presence of nuclear staining” [22]. Our study also suggests that the cases showing the presence of heterogeneous cytoplasmic staining with or without nuclear staining should be carefully interpreted.
Overall, the evidence from the publications presented in Table 1 and from the current study suggests that the cases should be scored as positive for BRAF V600E mutation if they display unequivocal, diffuse, uniform, granular, cytoplasmic staining in the majority of tumor cells at stain intensity ≥1. They should be scored as negative for BRAF V600E mutation if they exhibit no staining or weak, cytoplasmic, non-granular, non-uniform staining (stain intensity <1). The cases with staining of isolated tumor cells in a tumor that otherwise showed no staining should be considered negative. The cases should be considered as equivocal if they display ambiguous, heterogeneous, cytoplasmic staining with or without nuclear staining in tumor cells. If these interpretation criteria are followed the IHC with anti-BRAF V600E (VE1) antibody using recommended protocol with OptiView detection is optimal for detection of BRAF V600E mutation in CRC. In our study all 30 cases with BRAF V600E mutation showed unequivocal positive cytoplasmic staining in 85–100% tumor cells; all 30 cases with wild-type KRAS and BRAF were negative; 6.7% (4/60) cases with KRAS mutation showed heterogeneous, cytoplasmic/nuclear staining at stain intensity 1. However, the staining was heterogeneous and the presence of distinct nuclear staining was noted in these four cases along with cytoplasmic staining. Therefore, these cases were assigned as equivocal for BRAF V600E mutation. These cases were sequenced and confirmed to be negative for BRAF V600E mutation.
In summary, this study indicates that IHC with the anti-BRAF V600E (VE1) antibody performed on the Benchmark ULTRA automated stainer is a highly sensitive and specific detection method for determination of BRAF V600E mutation status in CRC. The results presented in this study are consistent with previous reports indicating that KRAS and BRAF V600E mutation are mutually exclusive. Based on our findings and consistent with other literature reports, the majority of BRAF V600E positive cases demonstrate a uniform or nearly uniform, diffuse staining pattern present in the majority of tumor cells. We propose that in the minority of cases with an equivocal staining pattern, additional molecular testing should be done to assess BRAF mutational status.
References
Adackapara CA, Sholl LM, Barletta JA et al (2013) Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 63:187–193
Affolter K, Samowitz W, Tripp S et al (2013) BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes, chromosomes & cancer 52:748–752
Bledsoe JR, Kamionek M, Mino-Kenudson M (2014) BRAF V600E immunohistochemistry is reliable in primary and metastatic colorectal carcinoma regardless of treatment status and shows high intratumoral homogeneity. Am J Surg Pathol 38:1418–1428
Boulagnon C, Dudez O, Beaudoux O et al (2016) BRAFV600E Gene Mutation in Colonic Adenocarcinomas. Immunohistochemical Detection Using Tissue Microarray and Clinicopathologic Characteristics: An 86 Case Series. Appl Immunohistochem Mol Morphol 24:88–96
Capper D, Preusser M, Habel A et al (2011) Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol 122:11–19
Capper D, Berghoff AS, Magerle M et al (2012) Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol 123:223–233
Capper D, Voigt A, Bozukova G et al (2013) BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. International journal of cancer Journal international du cancer 133:1624–1630
Davies H, Bignell GR, Cox C et al (2002) Mutations of the BRAF gene in human cancer. Nature 417:949–954
Day F, Muranyi A, Singh S et al (2015) A mutant BRAF V600E-specific immunohistochemical assay: correlation with molecular mutation status and clinical outcome in colorectal cancer. Target Oncol 10:99–109
Douillard JY, Oliner KS, Siena S et al (2013) Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 369:1023–1034
Dvorak K, Aggeler B, Palting J et al (2014) Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma. Pathology 46:509–517
Estrella JS, Tetzlaff MT, Bassett RL Jr et al (2015) Assessment of BRAF V600E Status in Colorectal Carcinoma: Tissue-Specific Discordances between Immunohistochemistry and Sequencing. Mol Cancer Ther 14:2887–2895
Hang JF, Li AF, Chang SC et al (2016) Immunohistochemical detection of the BRAF V600E mutant protein in colorectal cancers in Taiwan is highly concordant with the molecular test. Histopathology 69:54–62
Hartman DJ, Brand RE, Hu H et al (2013) Lynch syndrome-associated colorectal carcinoma: frequent involvement of the left colon and rectum and late-onset presentation supports a universal screening approach. Hum Pathol 44:2518–2528
Hernowo BS, Ariyanni F, Suryanti S et al (2014) Use of BRAF V600E as a molecular marker in aggressive colorectal cancer. Acta Med Indones 46:104–110
Ilie MI, Long-Mira E, Hofman V et al (2014) BRAFV600E mutation analysis by immunohistochemistry in patients with thoracic metastases from colorectal cancer. Pathology 46:311–315
Jin M, Hampel H, Zhou X et al (2013) BRAF V600E mutation analysis simplifies the testing algorithm for Lynch syndrome. Am J Clin Pathol 140:177–183
Koinuma K, Shitoh K, Miyakura Y et al (2004) Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer 108:237–242
Kuan SF, Navina S, Cressman KL et al (2014) Immunohistochemical detection of BRAF V600E mutant protein using the VE1 antibody in colorectal carcinoma is highly concordant with molecular testing but requires rigorous antibody optimization. Hum Pathol 45:464–472
Lasota J, Kowalik A, Wasag B et al (2014) Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the imunohistochemical approach. Am J Surg Pathol 38:1235–1241
Loes IM, Immervoll H, Angelsen JH et al (2015) Performance comparison of three BRAF V600E detection methods in malignant melanoma and colorectal cancer specimens. Tumour Biol 36:1003–1013
Marin C, Beauchet A, Capper D et al (2013) Detection of BRAF p.V600E Mutations in Melanoma by Immunohistochemistry Has a Good Interobserver Reproducibility. Arch Pathol Lab Med 138:71–75
Morkel M, Riemer P, Blaker H et al (2015) Similar but different: distinct roles for KRAS and BRAF oncogenes in colorectal cancer development and therapy resistance. Oncotarget 6:20785–20800
Nolan S, Arnason T, Drucker A et al (2014) The utility of BRAFV600E mutation-specific antibody for colon cancers with microsatellite instability. Appl Immunohistochem Mol Morphol 22:e8–e13
Oikonomou E, Koustas E, Goulielmaki M et al (2014) BRAF vs RAS oncogenes: are mutations of the same pathway equal? Differential signalling and therapeutic implications. Oncotarget 5:11752–11777
Pakneshan S, Salajegheh A, Smith RA et al (2013) Clinicopathological relevance of BRAF mutations in human cancer. Pathology 45:346–356
Parsons MT, Buchanan DD, Thompson B et al (2012) Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet 49:151–157
Piton N, Borrini F, Bolognese A et al (2015) KRAS and BRAF Mutation Detection: Is Immunohistochemistry a Possible Alternative to Molecular Biology in Colorectal Cancer? Gastroenterol Res Pract 2015:753903
Qiu T, Lu H, Guo L et al (2015) Detection of BRAF mutation in Chinese tumor patients using a highly sensitive antibody immunohistochemistry assay. Sci Rep 5:9211
Rajagopalan H, Bardelli A, Lengauer C et al (2002) Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418:934
Ren J, Li G, Ge J et al (2012) Is K-ras gene mutation a prognostic factor for colorectal cancer: a systematic review and meta-analysis. Dis Colon rectum 55:913–923
Ritterhouse LL, Barletta JA (2015) BRAF V600E mutation-specific antibody: A review. Semin Diagn Pathol 32:400–408
Rossle M, Sigg M, Ruschoff JH et al (2013) Ultra-deep sequencing confirms immunohistochemistry as a highly sensitive and specific method for detecting BRAF V600E mutations in colorectal carcinoma. Virchows Arch 463:623–631
Roth RM, Hampel H, Arnold CA et al (2015) A modified Lynch syndrome screening algorithm in colon cancer: BRAF immunohistochemistry is efficacious and cost beneficial. Am J Clin Pathol 143:336–343
Routhier CA, Mochel MC, Lynch K et al (2013) Comparison of 2 monoclonal antibodies for immunohistochemical detection of BRAF V600E mutation in malignant melanoma, pulmonary carcinoma, gastrointestinal carcinoma, thyroid carcinoma, and gliomas. Hum Pathol 44:2563–2570
Sajanti S, Sirnio P, Vayrynen JP et al (2014) VE1 immunohistochemistry accurately detects BRAF V600E mutations in colorectal carcinoma and can be utilized in the detection of poorly differentiated colorectal serrated adenocarcinoma. Virchows Arch 464:637–643
Schafroth C, Galvan JA, Centeno I et al (2015) VE1 immunohistochemistry predicts BRAF V600E mutation status and clinical outcome in colorectal cancer. Oncotarget 6:41453–41463
Seppala TT, Bohm JP, Friman M et al (2015) Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer 112:1966–1975
Sinicrope FA, Smyrk TC, Tougeron D et al (2013) Mutation-specific antibody detects mutant BRAF protein expression in human colon carcinomas. Cancer 119(15):2765–2770
Taieb J, Le Malicot K, Shi Q et al (2017) Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J Natl Cancer Inst 109. https://doi.org/10.1093/jnci/djw272
Thiel A, Heinonen M, Kantonen J et al (2013) BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Archiv : an international journal of pathology 463:613–621
Thiel A, Heinonen M, Kantonen J et al (2013) BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch 463:613–621
Toon CW, Walsh MD, Chou A et al (2013) BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol 37:1592–1602
Toon CW, Chou A, DeSilva K et al (2014) BRAFV600E immunohistochemistry in conjunction with mismatch repair status predicts survival in patients with colorectal cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 27:644–650
Vakiani E, Yaeger R, Brooke S et al (2015) Immunohistochemical detection of the BRAF V600E mutant protein in colorectal neoplasms. Appl Immunohistochem Mol Morphol 23:438–443
Yuan L, Chi Y, Chen W et al (2015) Immunohistochemistry and microsatellite instability analysis in molecular subtyping of colorectal carcinoma based on mismatch repair competency. Int J Clin Exp Med 8:20988–21000
Acknowledgements
The authors would like to thank Drs. Eric Walk, Mike Farrell and Stephen Billington for their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors are employees of Roche Tissue Diagnostics.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dvorak, K., Higgins, A., Palting, J. et al. Immunohistochemistry with Anti-BRAF V600E (VE1) Mouse Monoclonal Antibody is a Sensitive Method for Detection of the BRAF V600E Mutation in Colon Cancer: Evaluation of 120 Cases with and without KRAS Mutation and Literature Review. Pathol. Oncol. Res. 25, 349–359 (2019). https://doi.org/10.1007/s12253-017-0344-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0344-x