Abstract
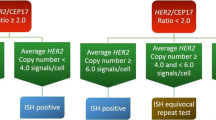
Breast cancer is the second leading cause of cancer mortality amongst American women. The HER2 gene encodes a cell surface receptor that affects cell proliferation and has been recognized as a diagnostic factor in treatment selection for invasive breast cancer. Examine accuracy in HER2 detection between manual count, computer assisted, and automated tiling algorithm. 42 randomly selected invasive breast cancer specimens were enumerated by fluorescence in situ hybridization (FISH)for HER2 and CEP17 markers using the Vysis HER2 assay (AbbotLaboratory, North Chicago, IL). Specimens were tested using three methods: Manual, computer assisted nuclei selection (Tissue FISH MetaSystems, Newton, MA), and automated enumeration (MetaSystems, Newton, MA). The greatest bias and widest agreement limits for HER2 and CEP17 were seen in Automatic versus Manual, the gold standard. HER2 values greater than 6 possessed the greatest bias and widest agreement limits. CEP17 comparison showed similar bias and agreement limits for each comparison. Kappa values indicated good agreement for all methods although Tissue FISH and Manual possessed better agreement. Higher agreement at lower HER2 & CEP17 count maybe due to fewer chromosomal aberrations, in which selection of field of views has less variation between methods. Alternatively, increased background signals seen in polyploidy may be responsible for the variations in signal count. Manual and Tissue FISH demonstrated good agreement amongst by both Altman Bland and Cohen’s Kappa. While the automatic method has good agreement at lower HER2, the sharp increase in variability at higher HER2 counts illustrates a limitation of the automatic method.


Similar content being viewed by others
References
Humphrey LL, Helfand M, Chan BK, Woolf SH (2002) Breast cancer screening: a summary of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med 137(5 Part 1):347–360
Heim S, Mitelman F. (2011) Cancer cytogenetics: chromosomal and molecular genetic abberations of tumor cells: John Wiley & Sons
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol Off J Am Soc Clin Oncol 31(31):3997–4013. doi:10.1200/JCO.2013.50.9984
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. (2005) Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 353(16):1659–1672
Hanna WM, Ruschoff J, Bilous M, Coudry RA, Dowsett M, Osamura RY, et al. (2014) HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol: an official journal of the United States and Canadian Academy of Pathology, Inc. 27(1):4–18. doi:10.1038/modpathol.2013.103
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235(4785):177–182
Hicks DG, Tubbs RR. (2005) Assessment of the HER2 status in breast cancer by fluorescence in situ hybridization: a technical review with interpretive guidelines. Hum Pathol 36(3):250–261. doi:10.1016/j.humpath.2004.11.010.
Furrer D, Jacob S, Caron C, Sanschagrin F, Provencher L, Diorio C. (2013) Validation of a new classifier for the automated analysis of the human epidermal growth factor receptor 2 (HER2) gene amplification in breast cancer specimens. Diagn Pathol 8:17. doi:10.1186/1746-1596-8-17
Myles P, Cui J. I. (2007) Using the bland–Altman method to measure agreement with repeated measures. Br J Anaesth 99(3):309–311.
Bland JM, Altman D. (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327(8476):307–310.
Viera AJ, Garrett JM. (2005) Understanding interobserver agreement: the kappa statistic. Fam Med 37(5):360–363.
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138(2):241–256. doi:10.5858/arpa.2013-0953-SA
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Thakral, G., Wey, A., Rahman, M. et al. Agreement of Different Methods for Tissue Based Detection of HER2 Signal in Invasive Breast Cancer. Pathol. Oncol. Res. 23, 79–84 (2017). https://doi.org/10.1007/s12253-016-0091-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-016-0091-4