Abstract
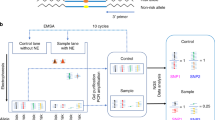
This study aims to explore the relationship between gene polymorphism and breast cancer, and to screen DEGs (differentially expressed genes) with SNPs (single nucleotide polymorphisms) related to breast cancer. The SNPs of 17 patients and the preprocessed SNP profiling GSE 32258 (38 cases of normal breast cells) were combined to identify their correlation with breast cancer using chi-square test. The gene expression profiling batch8_9 (38 cases of patients and 8 cases of normal tissue) was preprocessed with limma package, and the DEGs were filtered out. Then fisher’s method was applied to integrate DEGs and SNPs associated with breast cancer. With NetBox software, TRED (Transcriptional Regulatory Element Database) and UCSC (University of California Santa Cruz) database, genes-associated network and transcriptional regulatory network were constructed using cytoscape software. Further, GO (Gene Ontology) and KEGG analyses were performed for genes in the networks by using siggenes. In total, 332 DEGs were identified. There were 160 breast cancer-related SNPs related to 106 genes of gene expression profiling (19 were significant DEGs). Finally, 11co-correlated DEGs were selected. In genes-associated network, 9 significant DEGs were correlated to 23 LINKER genes while, in transcriptional regulatory network, E2F1 had regulatory relationships with 7 DEGs including MTUS1, CD44, CCNB1 and CCND2. KRAS with SNP locus of rs1137282 was involved in 35 KEGG pathways. The genes of MTUS1, CD44, CCNB1, CCND2 and KRAS with specific SNP loci may be used as biomarkers for diagnosis of breast cancer. Besides, E2F1 was recognized as the transcription factor of 7 DEGs including MTUS1, CD44, CCNB1 and CCND2.


Similar content being viewed by others
References
Sariego J (2010) Breast cancer in the young patient. Am Surg 76(12):1397–1400
Boyle P, Levin B (2008) World Cancer Report 2008. IARC Press, International Agency for Research on Cancer, Lyon
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64(1):9–29
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 100(7):3983
Sant M, Allemani C, Capocaccia R, Hakulinen T, Aareleid T, Coebergh JW, Coleman MP, Grosclaude P, Martinez C, Bell J (2003) Stage at diagnosis is a key explanation of differences in breast cancer survival across Europe. Int J Cancer 106(3):416–422
Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42(6):504–507
Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447(7148):1087–1093
Zardawi SJ, Zardawi I, McNeil CM, Millar EK, McLeod D, Morey AL, Crea P, Murphy NC, Pinese M, Lopez‐Knowles E (2010) High Notch1 protein expression is an early event in breast cancer development and is associated with the HER‐2 molecular subtype. Histopathology 56(3):286–296
Veronesi A, de Giacomi C, Magri MD, Lombardi D, Zanetti M, Scuderi C, Dolcetti R, Viel A, Crivellari D, Bidoli E (2005) Familial breast cancer: characteristics and outcome of BRCA 1–2 positive and negative cases. BMC Cancer 5(1):70
Satorra A, Bentler PM (2001) A scaled difference chi-square test statistic for moment structure analysis. Psychometrika 66(4):507–514
Sanges R, Cordero F, Calogero RA (2007) oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics 23(24):3406–3408
Whitlock M (2005) Combining probability from independent tests: the weighted Z‐method is superior to Fisher’s approach. J Evol Biol 18(5):1368–1373
Kim H-Y, Byun M-J, Kim H (2011) A replication study of genome-wide CNV association for hepatic biomarkers identifies nine genes associated with liver function. Biochem Mol Biol Rep 44(9):578–583
Yang J, Chen L, Wang L, Zhang W, Liu T, Jin Q (2007) TrED: the Trichophyton rubrum expression database. BMC Genomics 8(1):250
Wang Y, Wang DD (2013) University of California, Santa Cruz
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Nakaya A, Katayama T, Itoh M, Hiranuka K, Kawashima S, Moriya Y, Okuda S, Tanaka M, Tokimatsu T, Yamanishi Y (2013) KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res 41(D1):D353–D357
Schwender H, Krause A, Ickstadt K (2006) Identifying interesting genes with siggenes. The Newsletter of the R Project Volume 6/5, December 2006 34:45
Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H (2010) Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 28(10):1684–1691
Camphausen F, Kohl M, Ruckdeschel P, Stabla T, Ruckdeschel MP (2007) The distr Package
Irizarry RA, Hobbs B, Collin F, Beazer‐Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264
Tavassoli FA, Devilee P (2003) Pathology and genetics of tumours of the breast and female genital organs, vol 4. World Health Organization
Zuern C, Heimrich J, Kaufmann R, Richter KK, Settmacher U, Wanner C, Galle J, Seibold S (2010) Down-regulation of MTUS1 in human colon tumors. Oncol Rep 23(1):183–189
Frank B, Bermejo JL, Hemminki K, Sutter C, Wappenschmidt B, Meindl A, Kiechle-Bahat M, Bugert P, Schmutzler RK, Bartram CR (2007) Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis 28(7):1442–1445
Ye H, Pungpravat N, Huang B-L, Muzio LL, Mariggiò MA, Chen Z, Wong DT, Zhou X (2007) Genomic assessments of the frequent loss of heterozygosity region on 8p21. 3 ∼ p22 in head and neck squamous cell carcinoma. Cancer Genet Cytogenet 176(2):100–106
Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, Nicolas A, Chaverot N, Molinie V, Reyal F (2009) 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One 4(10):e7239
Bajorath J (2000) Molecular organization, structural features, and ligand binding characteristics of CD44, a highly variable cell surface glycoprotein with multiple functions. Proteins Struct Funct Bioinforma 39(2):103–111
Mayer B, Jauch K, Schildberg F, Funke I, Günthert U, Figdor C, Johnson J (1993) De-novo expression of CD44 and survival in gastric cancer. Lancet 342(8878):1019–1022
Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y (2008) CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 14(21):6751–6760
Alvero AB, Chen R, Fu H-H, Montagna M, Schwartz PE, Rutherford T, Silasi D-A, Steffensen KD, Waldstrom M, Visintin I (2009) Molecular phenotyping of human ovarian cancer stem cells unravel the mechanisms for repair and chemo-resistance. Cell Cycle (Georgetown, Tex) 8(1):158
Herrera-Gayol A, Jothy S (1999) Adhesion proteins in the biology of breast cancer: contribution of CD44. Exp Mol Pathol 66(2):149–156
Li Y, Chen Y-L, Xie Y-T, Zheng L-Y, Han J-Y, Wang H, Tian X-X, Fang W-G (2013) Association study of germline variants in CCNB1 and CDK1 with breast cancer susceptibility, progression, and survival among Chinese Han women. PLoS One 8(12):e84489
French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, Khan S, Maranian MJ, O’Reilly M, Hillman KM (2013) Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet 92(4):489–503
Song H, Hogdall E, Ramus SJ, DiCioccio RA, Hogdall C, Quaye L, McGuire V, Whittemore AS, Shah M, Greenberg D (2008) Effects of common germ-line genetic variation in cell cycle genes on ovarian cancer survival. Clin Cancer Res 14(4):1090–1095
Driver KE, Song H, Lesueur F, Ahmed S, Barbosa-Morais NL, Tyrer JP, Ponder BA, Easton DF, Pharoah PD, Dunning AM (2008) Association of single-nucleotide polymorphisms in the cell cycle genes with breast cancer in the British population. Carcinogenesis 29(2):333–341
Johnson DG (2000) The paradox of E2F1: oncogene and tumor suppressor gene. Mol Carcinog 27(3):151–157
Abdraboh ME, Gaur RL, Hollenbach AD, Sandquist D, Raj MH, Ouhtit A (2011) Survivin is a novel target of CD44-promoted breast tumor invasion. Am J Pathol 179(2):555–563
Huang L (2008) Carrot and stick: HIF-α engages c-Myc in hypoxic adaptation. Cell Death Differ 15(4):672–677
Wang WY, Chien YC, Wong YK, Lin YL, Lin JC (2012) Effects of KRAS mutation and polymorphism on the risk and prognosis of oral squamous cell carcinoma. Head Neck 34(5):663–666
Yen L-C, Yeh Y-S, Chen C-W, Wang H-M, Tsai H-L, Lu C-Y, Chang Y-T, Chu K-S, Lin S-R, Wang J-Y (2009) Detection of KRAS oncogene in peripheral blood as a predictor of the response to cetuximab plus chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res 15(13):4508–4513
Acknowledgments
This study was supported by Science and technology innovation projects in henan province department of education (NO. 4206).
Conflict of Interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
Highlights
1 Eleven significant DEGs were identified in breast cancer.
2 MTUS1, CD44, CCNB1, CCND2 and KRAS with specific SNPs were vital genes for breast cancer.
3 E2F1 was identified as the transcription factor of MTUS1, CD44, CCNB1 and CCND2.
4 Significant DEGs enriched in multiple cancer signaling pathways.
Rights and permissions
About this article
Cite this article
Yuan, P., Liu, D., Deng, M. et al. Identification of Differently Expressed Genes with Specific SNP Loci for Breast Cancer by the Integration of SNP and Gene Expression Profiling Analyses. Pathol. Oncol. Res. 21, 469–475 (2015). https://doi.org/10.1007/s12253-014-9851-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-014-9851-1