Abstract
African swine fever virus (ASFV), as a member of the large DNA viruses, may regulate autophagy and apoptosis by inhibiting programmed cell death. However, the function of ASFV proteins has not been fully elucidated, especially the role of autophagy in ASFV infection. One of three Pyrroline-5-carboxylate reductases (PYCR), is primarily involved in conversion of glutamate to proline. Previous studies have shown that depletion of PYCR2 was related to the induction of autophagy. In the present study, we found for the first time that ASFV E199L protein induced a complete autophagy process in Vero and HEK-293T cells. Through co-immunoprecipitation coupled with mass spectrometry (CoIP-MS) analysis, we firstly identified that E199L interact with PYCR2 in vitro. Importantly, our work provides evidence that E199L down-regulated the expression of PYCR2, resulting in autophagy activation. Overall, our results demonstrate that ASFV E199L protein induces complete autophagy through interaction with PYCR2 and down-regulate the expression level of PYCR2, which provide a valuable reference for the role of autophagy during ASFV infection and contribute to the functional clues of PYCR2.
Similar content being viewed by others
Introduction
African swine fever (ASF) is a fatal disease caused by African swine fever virus (ASFV), leading to severe hemorrhagic fever in pigs. In 1920s, ASFV was first found in Kenya and has continued to spread worldwide since then (Arzt et al. 2010). The first ASF case broke out in China on August 3, 2018 in Liaoning and rapidly spread across the entire country (Zhao et al. 2019). ASFV has seriously threatened the world's pig industry due to the lack of vaccines. To promote vaccine development, improved knowledge is needed of how the virus interacts with the host and the mechanism of antiviral defense. The first and critical step is to explain the function of its viral protein.
The ASFV viral particle displays a unique icosahedral morphology with a diameter of about 250 nm (Wang et al. 2019). The double-stranded DNA genome of the virus codes for more than 150 proteins of which many are of unknown function, including E199L. As early as 1996, researchers identified the protein E199L, also named j18L, which was expressed late post-infection and localized in the viral factories (Sun et al. 1996). But no specific functional studies have been reported until 2020. Matamoros addressed the role of ASFV protein E199L in virus replication and results showed that E199L is required for viral core entry but not for virus morphogenesis due to its inner envelope transmembrane polypeptide structure containing intramolecular disulfide bonds (Matamoros et al. 2020).
Autophagy is a highly conserved catabolic process that is routinely described as a common strategy for cell protection. Autophagosomes fuse with lysosomes forming autolysosomes, resulting in the degradation of the engulfed components and generating nutrients (Campbell et al. 2018). Lysosomal degradation mediated by autophagy enhances the immune response to HIV infection in a variety of ways (Nardacci et al. 2017). However, many viruses have evolved various methods to escape autophagic degradation (Choi et al. 2018). In HIV-infected macrophages, NEF binds to Beclin-1 to block the degradative stages of autophagy and the accumulation of autophagic factors (Kyei et al. 2009). Further on, some viruses, like Zika virus (Peng et al. 2018) and other flavivirus (McLean et al. 2011), even can hijack cellular autophagy to benefit viral replication. This may because the double-membrane compartment formed during autophagy provides a physical platform for viral replication and protects viral RNA from detection and degradation by innate immune sensors (Choi et al. 2018).
However, the research between autophagy and ASFV infection is still insufficient. An independent study found that ASFV inhibited autophagy probably because its A179L protein interacts with Beclin-1 to control autophagy (Hernaez et al. 2013). The latest study reported the structural basis for A179L-mediated inhibition of autophagy may be by binding to the Beclin-1 BH3 motif (Banjara et al. 2019). In addition, ASFV infection did not appear to induce LC3 lipidation or autophagosome formation in Vero cells. What is interesting, however, is that induction of autophagy by rapamycin prior to ASFV infection reduces viral infectivity (Alonso et al. 2013). So, more relevant research is needed urgently. In general, the role of autophagy in the process of ASFV infection remains unclear and further studies are needed.
Pyrroline-5-carboxylate reductase 2 (PYCR2), one of three Pyrroline-5-carboxylate reductases, is primarily involved in the conversion of glutamate to proline (De Ingeniis et al. 2012). But over the years, few studies on PYCR2 have been reported. Until 2015, Nakayama showed that mutations in PYCR2 cause microcephaly and hypomyelination (Nakayama et al. 2015). More research later confirmed that PYCR2 deficiency is associated with a rare autosomal recessive genetic disease (Zaki et al. 2016; Afroze et al. 2020). In addition, PYCR2 collaborates with RRM2B to protect cells for overt oxidative stress (Kuo et al. 2016). Likewise, another study also showed that PYCR2 has significant antioxidant activity and can inhibit the upregulation of ROS levels (Nakayama et al. 2015). Recent research has shown that loss of PYCR2 causes neurodegeneration by increasing cerebral glycine levels via SHMT2 (Escande-Beillard et al. 2020). It is noteworthy that down-regulation of PYCR2 induces autophagy in melanoma cells (Ou et al. 2016). This result surprised us by showing that there may be some association between PYCR2 and autophagy.
In the present study, transient transfection of recombinant plasmid expressing ASFV-E199L protein could induce a complete autophagy process in Vero cells and human embryonic kidney-293T (HEK-293T) cells. Based on Co-immunoprecipitation coupled with mass spectrometry (CoIP-MS) analysis, we identified 119 potential E199L-interacting proteins in Vero cell line. After in vitro validation of E199L/PYCR2 binding, we investigated the functional meaning of this novel interaction. Our data indicate that E199L can down-regulate the expression level of PYCR2 and ultimately promote cellular autophagy. In general, to an extent, our research has filled a gap in the study on African swine fever virion proteins function. Moreover, we also supplemented the validation of the relationship between PYCR2 and autophagy.
Materials and Methods
Cell Cultures and Transfection
Vero cells and HEK-293T cells were maintained at 37 °C in complete Dulbecco’s modified Eagle’s medium (C11995500BT, Thermo Fisher) containing 10% fetal bovine serum (10,099–141, Gibco). Small interfering RNA (siRNA) targeting PYCR2(5′-UUCAUGGCUCUGGACGCAUUGTT-3′) and the negative control (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by Sangon Biotech. Cells grown to approximately 80% confluence were transfected with plasmid or siRNA by using Lipofectamine® 3000 Reagent (L3000015, Invitrogen).
Plasmid Constructions
pPRK5-Flag, pEGFP-C1 and pmCherry-N1 were preserved in laboratory. pPRK5-Flag-E199L and pEGFP-E199L were constructed by synthetizing ASFV E199L gene into pPRK5-Flag and pEGFP-C1 vector. pPRK5-Flag-E183L, pPRK5-Flag-F317L, pPRK5-Flag-MGF 505-4R, pPRK5-Flag-CP530R and pPRK5-Flag-B602L were constructed by synthetizing ASFV E183L/F317L/MGF 505-4R/CP530R/B602L gene into pPRK5-Flag vector. pPRK5-Flag-PYCR2 and pmCherry-PYCR2 was constructed by cloning pig PYCR2 (NCBI Reference Sequence: XM_003130542.4) into pPRK5-Flag and pmCherry-N1 vector. pmCherry-ARL8B, pmCherry-BAG5, pmCherry-FLII and pmCherry-LAMP2 was constructed by cloning pig ARL8B (NM_001252214.1)/BAG5 (XM_021081511.1)/FLII (XM_021067982.1)/LAMP2 (NM_001244255.1) into pmCherry-N1 vector. The method of homologous recombination was used to construct these plasmids and confirmed by sequencing.
Antibodies and Reagents
The antibodies used in the study are as follows: Anti-LC3B (L7543) and anti-SQSTM1 (P0067) antibodies were obtained from Sigma-Aldrich. Anti-Flag (14793S) and anti-β-actin (4970S) were obtained from Cell Signaling Technology. Anti-PYCR2 (17,146–1-AP), horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit secondary antibodies (SA00001), coralite 594/488 conjugated goat anti-mouse/rabbit IgG(H + L) antibodies (SA00013) were obtained from Protein Tech Group. Anti-mCherry (183,628) antibody was purchased from abcam and anti-GFP (T0005) antibody was purchased from affinity. PAGE Gel Fast Preparation Kit (PG112 and PG114), Protein Free Rapid Blocking Buffer (PS108) and Omni-ECL™ Enhanced Pico Light Chemiluminescence Kit (SQ101) were obtained from Epi Zyme. Electrophoresis buffer (FD9001) and Western transfer buffer (FD9002) were obtained from Fdbio science.
Reverse Transcription-Quantitative PCR
Total RNA was extracted from cultured cells by using Trizol reagent and 1 μg RNA was reverse-transcribed using Prime Script RT Reagent Kit (AK2601, Takara Bio). The qRT-PCR was performed with DyNAmo Flash SYBR Green qPCR Kit (F415XL, Thermo Fisher). The primer sequences for PYCR2 were 5′-CCTGCATCCGCACAAGAGAG-3′ (forward) and 5′-GTGGGGGACTCCAGTTTCACT-3′ (reverse). GAPDH was used as the internal control and its sequences were: 5′- ATCCCGCCAACATCAAATGG -3′ (forward) and 5′- ACTTCTCATGGTTCACGCCC -3′ (reverse). Relative expression of PYCR2 was normalized and the data were analyzed using the 2−ΔΔCt method.
Western Blot
Prepared cells were dissolved in 100 µL RIPA lysis buffer (CW2333S, CWBIO) including 100× protease inhibitor (CW2200, CWBIO). After centrifugation, the lysates were denatured for 5 min in 5× SDS-PAGE loading buffer (P0015, Beyotime). Proteins were separated on an SDS-PAGE gel and transfered to a PVDF membrane. Next, we block it with 5% skimmed milk powder at 4 °C overnight. After that, the membrane was incubated with primary antibodies for 1 h, then the corresponding secondary antibodies at room temperature for 0.5 h. Developed membranes were imaged using an Azure c300 digital imager system (Azure Biosystems, Dublin, CA).
Immunofluorescence and Confocal Microscopy
Vero and HEK-293T Cell monolayers were fixed using 4% paraformaldehyde (AAPR12-500, Python bio) for 15 min at room temperature. Then, cells were permeabilized with 0.1% Triton-X 100 (AAPR96-C100, Python bio) for 15 min at room temperature. Next, we block it with 5% skimmed milk powder at 4 °C overnight. After that, cells were incubated with primary antibodies for 1 h at room temperature, then the fluorochrome-conjugated secondary antibodies for 1 h in the dark. Last, cells were incubated with DAPI (P36941, Invitrogen) for nuclear staining. The fluorescence signals were detected with a TCS SP8 confocal fluorescence microscope (Leica).
Co-Immunoprecipitation
After transfection with the indicated plasmids, cells were lysed in ice-cold lysis buffer. Lysates were centrifuged at 12,000 ×g for 10 min and then supernatants were incubated with GFP-Trap agarose beads (gta-20, Chromo Tek) for 1 h at 4 °C. After washing 3 times by wash buffer, beads were boiled with 2× SDS loading buffer. Proteins of interest were analyzed by western blotting.
Mass Spectrometry Analysis
Mass spectrometry analysis was performed at Cloud-Seq Biotech Ltd. Co. (Shanghai, China). FASP method is mainly used for enzymatic hydrolysis and the specific steps are as follows (Wisniewski et al. 2009). Each sample was treated with 500 μL UA buffer (8 mol/L Urea, 150 mmol/L Tris–HCl, pH 8.0) after treatment in a 10 kDa ultrafiltration centrifuge tube at 5000 ×g at 15 °C. 500 μL UA buffer was added and mixed with DTT until the final concentration was 100 mmol/L. The mixture was centrifuged at 8000 ×g for 15 min. Then 500 μL UA buffer was added and centrifuged at 8000 ×g for 15 min to discard the filtrate. 500 μL IAA (50 mmol/L IAA in UA) was added, which was oscillated at 600 rpm for 1 min, and incubate at room temperature for 30 min in darkness and centrifuge at 8000 ×g for 10 min. 500 μL UA buffer was added and centrifuged at 8000 ×g for 10 min and repeat twice. Then 4 μL trypsin buffer (2 μg trypsin) and 500 μL NH4HCO3 were added, which was oscillated at 600 rpm for 1 min and incubate 16 h at 37 °C. Finally, the filtrate was collected by centrifugation at 14,000 ×g for 10 min and completely dried down. Gradient elution was performed with mobile phase A (0.1% formic acid solution) and B (99% ACN/0.1% formic acid). Peptides were eluted with a gradient of 2%–6% B over 2 min, followed by 6%–30% B over 98 min, 30%–45% B over 9 min, 45%–90% B over 2 min and held at 90% B for 10 min.
Bioinformatics Analysis
The mass—charge ratio of peptides and fragments of peptides are collected as follows. After each full scan (MS1 scan), 12 fragments (MS2 scan) were collected. MS1 scan represented profile mode with a resolution of 70,000 and MS2 scan represented profile mode with a resolution of 17,500. Collision energy: 27.0%, isolation window: 1.2 m/z. Raw File used Max Quant 1.6.0.16 to analyze data (Cox et al. 2008), and Max Quant's built-in algorithm was used to identify polypeptides and proteins, and relative quantitative information (LFQ) was given (Cox et al. 2014). The LFQ value was converted by log10 and quantile standardized (LIMMA package), then the differential polypeptides and proteins were screened by the use of volcanic diagrams, and the clustering, scatter, and volcanic diagrams were drawn. The GO and KEGG pathways of related genes were analyzed.
Statistical Analysis
Western blot and Co-IP results are representatives of three independent experiments. All statistics were calculated using Prism 8 (Graph pad). The Student t test was used to evaluate the statistical significance and P < 0.05 was considered statistically significant.
Results
ASFV E199L Protein Triggers Autophagy in VERO and HEK-293T Cells
To investigate whether individual ASFV proteins trigger autophagy, we transfected Flag-tagged ASFV E183L, E199L, F317L, MGF 505-4R, CP530R and B602L expression plasmids into Vero cells and used Western blot to measure the conversion of LC3-I to LC3-II and P62 degradation, which is a general method for evaluating autophagy process (Bjorkoy et al. 2005). As shown in Fig. 1A, transient transfection of E199L protein induced the conversion of LC3-I to LC3-II and P62 degradation. We carried out densitometry analysis of the density band and the statistical results also confirmed the induction of autophagy (Fig. 1B). In contrast, E183L, F317L, CP530R and B602L proteins did not induce autophagy.
ASFV E199L protein triggers autophagy in Vero and HEK-293T cells. A Vero cells were transfected with empty vectors or various plasmids expressing Flag-tagged ASFV E183L, E199L, F317L, MGF 505-4R, CP530R, B602L proteins and empty vector (EV). At 24 h post-transfection, cells were harvested and Western blotting was performed. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. B Densitometric LC3-II/LC3-I and P62/β-actin ratios from at least 3 independent experiments were shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student's test. ***P < 0.001. C and E Vero and HEK-293T cells were transfected with ASFV E199L protein expression plasmids (E199L) or empty vectors (EV). Cells were harvested at indicated time points (6, 12, 24 and 36 h) and detected with anti-LC3B antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. D and F Densitometric LC3-II/LC3-I ratios from at least 3 independent experiments were shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student's test. *P < 0.05, **P < 0.01, ***P < 0.001. G and H Vero and HEK-293T cells were transfected with ASFV E199L protein expression plasmids or empty vectors. The fluorescent puncta of LC3B were observed by confocal microscopy with scale bars indicating 10 µm.
To confirm autophagy induction by ASFV E199L protein, we measured induction over time after transfection of ASFV E199L protein. The conversion of LC3-I to LC3-II was monitored at 6, 12, 24, and 36 h post-transfection with empty vector (EV) treatment as a negative control. As demonstrated in Fig. 1C, E199L-transfected Vero cells had significant conversion of LC3-I to LC3-II since 12 h. To quantify these results, we further used densitometry analysis to measure band intensity. The ratio of LC3-II to LC3-I was increased at 12, 24, and 36 h in ASFV E199L-transfected cells but not in untreated control cells at the corresponding time points (Fig. 1D). Accordingly, we detected similar phenomena in HEK-293T cells (Fig. 1E, 1F).
To further confirm that the conversion of LC3-I to LC3-II was indeed related to the induction of autophagy, we further observed the formation of LC3 fluorescent puncta from the morphological point of view. Vero and HEK-293T cells were transfected with pPRK5-Flag-E199L for 24 h. In ASFV E199L-transfected cells, the amount of LC3 fluorescent puncta noticeably increased while a largely dispersed fluorescence distribution was observed in EV-treated groups (Fig. 1G, 1H). Collectively, these data demonstrate that pPRK5-Flag-E199L transfection indeed induced an autophagic response in Vero and HEK-293T cells.
ASFV E199L Protein-Induced Autophagy Is a Complete Process
The accumulation of autophagosomes may be due to autophagy induction or a block in autophagosome maturation (Klionsky et al. 2016). To further explore whether the autophagic response triggered by E199L was a complete process, the expression level of P62 was measured by western blot analysis. As an adaptor of LC3-II, P62 was generally considered a marker to asses autophagic flux (Yasui et al. 2016). Compared to EV-treated group, P62 was not significantly degraded at early stages, but significantly decreased at later stages (24 and 36 h) (Fig. 2A, 2B), suggesting that the E199L protein induced complete autophagy. Furthermore, we observed the similar results in HEK-293T cells (Fig. 2C, 2D).
ASFV E199L protein-induced autophagy is a complete process. A and C Vero and HEK-293T cells were transfected with ASFV E199L protein expression plasmids or empty vectors. Cells were harvested at indicated time points (6, 12, 24 and 36 h) and detected with anti-P62 antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. B and D Densitometric P62/β-actin ratios from at least 3 independent experiments were shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student’s test. ***P < 0.001. E and F Vero and HEK-293T cells were transfected with ASFV E199L protein expression plasmids or empty vectors for 24 h and then treated with Lyso-Tracker for 2 h. The fusion between the autophagosomes and lysosomes were observed by confocal microscopy with scale bars indicating 10 µm.
Subsequently, to investigate the accumulation of autophagosomes, cells were labeled with Lyso-Tracker Red to label acidic compartments or organelles in living cells (Gu et al. 2019). Autophagosomes and Lyso-Tracker Red were co-localized in cells following E199L-transfected. By contrast, in EV-treated group, almost no co-localization was observed between Autophagosomes and Lyso-Tracker Red (Fig. 2E, 2F). These results suggested that autophagosomes could fuse with acidic compartments or organelles following the transfection of E199L. Taken together, these data indicate that E199L protein-induced autophagy is a complete process in Vero and HEK-293T cells.
ASFV E199L Protein Interacts with Host Protein PYCR2
The study of protein–protein interactions is an essential process to understand the biological functions of proteins and the underlying mechanisms. Co-immunoprecipitation coupled with mass spectrometry (CoIP-MS) is one of the most extensively used high-throughput techniques to discover novel protein–protein interactions (Ngounou et al. 2014). To explore this issue, we preformed mass spectrometry analysis of E199L protein and screened 119 potential interacting proteins (Supplementary Table S1). To identify putative functional processes associated with E199L-interacting proteins, we used GO cluster analysis to provide relevant information about biological processes (BP), cellular components (CC) and molecular functions (MF) and KEGG pathway analysis (Supplementary Fig. S1). We have noted that the GO term intracellular and membrane-bounded organelle were enriched in the CC category, while the binding-associated GO terms were enriched in the MF category, and various metabolic processes were enriched in the BP category. The top 10 KEGG pathways enriched by the target genes included ribosome, tight junction and endoplasmic reticulum pathways.
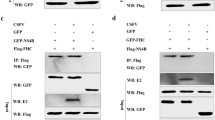
Furthermore, we have chosen 5 possible interaction proteins (ARL8B, LAMP2, PYCR2, BAG5 and FLII) which are related to cell autophagy and constructed eukaryotic expression plasmid respectively (Table 1). Co-immunoprecipitation assays and Western blot analysis showed that PYCR2 can interact with E199L (Fig. 3A) whereas others do not (Supplementary Fig. S2). In addition, confocal immunofluorescence suggested that host protein PYCR2 (mCherry) co-localized with ASFV-E199L (EGFP) in the cytoplasm (Fig. 3B). This further illustrates the interaction between PYCR2 and E199L. Interestingly enough, we found that the expression levels of PYCR2 seem to be regulated by E199L. After ASFV E199L-transfected, the fluorescence puncta of PYCR2 significantly decreased as shown in Fig. 3B.
ASFV E199L protein interacts with host protein PYCR2. A Vero cells were co-transfected with pEGFP-C1, pEGFP-E199L, pmCherry-N1 and pmCherry-PYCR2 plasmid in pairs and immunoprecipitation was performed with anti-EGFP antibody. Immunoblotting analysis was performed with anti-EGFP antibody and anti-mCherry antibody. Blots are representative of the 3 independent experiments. B Vero cells were co-transfected with pEGFP-C1, pEGFP-E199L, pmCherry-N1 and pmCherry-PYCR2 plasmid in pairs. The fluorescence signals were visualized by confocal immunofluorescence microscopy with scale bars indicating 10 µm. C Vero cells were transfected with ASFV E199L protein expression plasmids or empty vectors. Quantitative real-time PCR of the mRNA expression levels of PYCR2 were detected. GAPDH, as an inner reference, was used as sample-loading control. D Vero cells were transfected with ASFV E199L protein expression plasmids or empty vectors. At 24 h post-transfection, cells were harvested and western blotting was performed with anti-PYCR2 antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. Densitometric PYCR2/β-actin ratios from at least 3 independent experiments are shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student’s test. ***P < 0.001.
To further clarify the detailed interaction between E199L and PYCR2, the expression levels of PYCR2 were measured in E199L-transfected cells. As we expected, the overexpression of E199L down-regulated the expression of PYCR2 both in mRNA and protein levels (Fig. 3C, 3D). These data demonstrate that the ASFV E199L protein interacts with host protein PYCR2 and down-regulates the expression level of PYCR2.
Host Protein PYCR2 Regulates Autophagy Activation
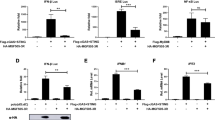
There is little known about the function of PYCR2 and only one literature suggests that the down-regulation of PYCR2 seems to be associated with autophagy (Ou et al. 2016). We therefore hypothesized that the E199L-induced autophagy was mediated by down-regulation of PYCR2. To put this hypothesis to the test, we knocked down the expression of PYCR2 and detected the changes of autophagy markers. First, we verified the interference efficiency of siRNA by Western blot analysis. As shown in Fig. 4A, 4B, siRNA-1 has the highest interference effect in 24 h. After that, we collected cellular proteins treated by siRNA-1 at different time points. In siRNA-treated cells, the conversion of LC3-I to LC3-II and the degradation of P62 were observed as against NC-treated cells. Densitometry analysis also supports the results (Fig. 4C, 4D). These data indicate that depletion of PYCR2 promotes autophagy and it is consistent with previous studies (Ou et al. 2016).
Host protein PYCR2 regulates autophagy activation. A and B Vero cells were transfected with si-PYCR2 and si-NC treatment was used as negative control. At 36 h post-transfection, cells were harvested and western blotting was performed with anti-PYCR2 antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. Densitometric PYCR2/β-actin ratios from at least 3 independent experiments were shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student’s test. ***P < 0.001. C and E Cells were harvested at indicated time points (24, 36 and 48 h) and detected with anti-P62 antibody and anti-LC3B antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. C: Vero cells were transfected with si-PYCR2 and si-NC. E: Vero cells were transfected with ASFV E199L protein expression plasmids or empty vectors. D and F Densitometric LC3-II/LC3-I and P62/β-actin ratios from at least 3 independent experiments are shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student’s test. ** P < 0.01, ***P < 0.001. ns:not significant.
To further investigate the regulation of PYCR2 on autophagy, we made use of a PYCR2 overexpression vector (pPRK5-Flag-PYCR2). We checked the influence of PYCR2 overexpression on autophagy as assessed by monitoring the conversion of LC3-I to LC3-II and the degradation of P62. Out of our expectation, overexpression of PYCR2 had no impact on these processes, which indicated the existence of complicated mechanism in this regulation (Fig. 4E, 4F).
ASFV E199L Protein Induces Autophagy by Down-Regulating PYCR2
In order to clarify the interaction between E199L and PYCR2 on autophagy, we transfected cells with multiple plasmids either individually or co-transfected. As shown in Fig. 5, overexpression of E199L or knockdown of PYCR2 promoted autophagy to varying degree, and when E199L was co-transfected with si-PYCR2, the results were the same. Nevertheless, when PYCR2 was overexpressed, the activity of P62 inhibited by E199L was completely recovered; and the level of LC3-II/LC3-I was decreased. These results further confirmed that E199L induce autophagy by interacting with PYCR2 and down-regulating the expression level of PYCR2.
ASFV E199L protein induces autophagy by down-regulating PYCR2. A and C Vero cells were transfected with various expression plasmids. At 24 h post-transfection, cells were harvested and western blotting was performed with anti-PYCR2 antibody. Blots are representative of the 3 independent experiments. β-actin was used as sample-loading control. B and D Densitometric LC3-II/LC3-I and P62/β-actin ratios from at least 3 independent experiments were shown. Error bars show standard error of the mean (SEM). Significance was analyzed with two-tailed Student’s test. **P < 0.01, ***P < 0.001.
Discussion
Within global range, ASFV leads to devastating effect on pig production (Costard et al. 2009). However, owing to complex virus structure and unknown protein function, it is difficult to make a breakthrough in vaccine development (Arias et al. 2017). In order to better explain the pathogenic mechanism of ASFV and promote vaccine development, it is an important step to reveal the unknown protein functions of ASFV. We first concentrate efforts on autophagy because it is closely related to viral infection. For viruses, cell autophagy is a double-edged sword. In general, autophagy is described as a strategy for cell protection. In part that is because autophagy can be utilized to degrade viral particles. Some viruses, however, can subvert this autophagic degradation and even help themselves survive by promoting cell autophagy (Choi et al. 2018). As is known to all, ASFV infection is a complex physiological process and whether autophagy is involved in it is still uncertain. In this study, we first confirmed that ASF viral protein E199L can induce autophagy in Vero and HEK-293T cells.
The occurrence of autophagy usually includes three indicators: the formation of LC3 fluorescent spot, the accumulation of LC3-II and the degradation of P62 (Zhang et al. 2016). Western blot analysis demonstrated an increase in the ratio of LC3-II/LC3-I band density at different time points (12, 24 and 36 h) after transfections with E199L eukaryotic expression plasmid. At the same time, through confocal microscopy, we observed the formation of LC3 fluorescent spots. In addition, the degradation of P62 was occurred starting at 24 h after E199L protein transfections. We also discovered the co-location of GFP-LC3 and Lyso-Tracker Red by confocal microscopy. These observations confirmed that ASF viral protein E199L promotes the fusion of autophagosomes with lysosomes and induces complete autophagy in Vero and HEK-293T cells.
To further investigate the function of E199L, we performed affinity purification-mass spectrometry analysis of E199Linteractome. A total 119 proteins were identified in FLAG-E199L immunocomplexes after subtracting the proteins found in the immunocomplexes control experiments with FLAG-empty vector. The 119 E199L interactors were assigned to 498 GO categories and the terms were summarized by the three main GO categories and 30 subcategories (Supplementary Fig. S1). For biological processes, most proteins were linked to various metabolic process. For cellular component, most proteins were associated with intracellular and membrane-bounded organelle. These results partly explain the functional basis of E199L for membrane fusion and core penetration (Matamoros et al. 2020). For molecular function, E199L interactors showed significant enrichment in binding-associated GO terms, such as DNA/RNA binding, protein binding and heterocyclic compound binding. According to the KEGG pathways analysis, E199L-interacting proteins were enriched in ribosome, tight junction and endoplasmic reticulum pathways. In conclusion, these results provide references for further study of the function of ASF-E199L protein.
Host protein pyrroline-5-carboxylate reductase 2 (PYCR2) was identified as target protein by Co-immunoprecipitation. Laser confocal experiments also verified the interaction between them. Interestingly, down-regulation of the PYCR2 fluorescence puncta was observed under confocal microscopy in E199L-overexpressed group. Moreover, we perform RT-qPCR experiments and western blot analysis to demonstrate that E199L can down-regulate the expression level of PYCR2 at mRNA and protein levels, respectively. To further confirm our hypothesis that E199L promotes autophagy by down-regulating PYCR2, we transfected cells with multiple plasmids either individually or co-transfected. These results of western blot experiments strongly support this view.
PYCR2 is primarily involved in conversion of glutamate to proline (De Ingeniis et al. 2012). There are few reports on its more extensive functional studies. But we note that the down-regulation of PYCR2 seems to be closely related to autophagy (Ou et al. 2016). We found that the depletion of PYCR2 promotes cellular autophagy, but overexpression had no significant impact on these processes. PYCR2 has been shown to have significant antioxidant activity, including inhibition of ROS levels, maintenance of mitochondrial function, and promotion of cancer cell survival (Nakayama et al. 2015; Kuo et al. 2016). This may explain the association between PYCR2 reduction and increased autophagy. Because the moderate increase of ROS leads to oxidoreduction imbalance, and autophagy is induced follow as (Yuan et al. 2018). However, inhibition of ROS levels does not inhibit autophagy in the same way. More importantly, whether the overexpression of PYCR2 can recover the autophagy level induced by oxidative stress imbalance deserves further study. In fact, in this study, E199L-induced autophagy could be recovered by overexpression of PYCR2.
Overall, our results confirmed that ASF viral protein E199L triggers autophagy by interacting with PYCR2 and down-regulates the expression level of PYCR2. These conclusions provide a valuable reference for studying the relationship between ASF viral proteins and autophagy, and also contribute to the functional clues of PYCR2.
References
Afroze B, Mercimek-Andrews S (2020) Pyrroline-5-Carboxylate Reductase 2 Deficiency: A New Case and Review of the Literature. Can J Neurol Sci 47:280–282
Alonso C, Galindo I, Cuesta-Geijo MA, Cabezas M, Hernaez B, Munoz-Moreno R (2013) African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res 173:42–57
Arias M, de la Torre A, Dixon L, Gallardo C, Jori F, Laddomada A, Martins C, Parkhouse RM, Revilla Y, Rodriguez FAJ (2017) Approaches and perspectives for development of African swine fever virus vaccines. Vaccines 5:35
Arzt J, White WR, Thomsen BV, Brown CC (2010) Agricultural diseases on the move early in the third millennium. Vet Pathol 47:15–27
Banjara S, Shimmon GL, Dixon LK, Netherton CL, Hinds MG, Kvansakul M (2019) Crystal structure of african swine fever virus A179L with the autophagy regulator beclin. Viruses 11:789
Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614
Campbell GR, Bruckman RS, Herns SD, Joshi S, Durden DL, Spector SA (2018) Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J Biol Chem 293:5808–5820
Choi Y, Bowman JW, Jung JU (2018) Autophagy during viral infection: a double-edged sword. Nat Rev Microbiol 16:341–354
Costard S, Wieland B, de Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK (2009) African swine fever: How can global spread be prevented? Philos Trans R Soc Lond B Biol Sci 364:2683–2696
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372
Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526
De Ingeniis J, Ratnikov B, Richardson AD, Scott DA, Aza-Blanc P, De SK, Kazanov M, Pellecchia M, Ronai Z, Osterman AL (2012) Functional specialization in proline biosynthesis of melanoma. PLoS One 7:e45190
De Snoo ML, Friesen EL, Zhang YT, Earnshaw R, Dorval G, Kapadia M, O’Hara DM, Agapova V, Chau H, Pellerito O, Tang MY, Wang X, Schmitt-Ulms G, Durcan TM, Fon EA, Kalia LV, Kalia SK (2019) Bcl-2-associated athanogene 5 (BAG5) regulates Parkin-dependent mitophagy and cell death. Cell Death Dis 10:907
Escande-Beillard N, Loh A, Saleem SN, Kanata K, Hashimoto Y, Altunoglu U, Metoska A, Grandjean J, Ng FM, Pomp O (2020) Loss of PYCR2 causes neurodegeneration by increasing cerebral glycine levels via SHMT2. Neuron 107:82–94
Fukushima M, Inoue T, Miyai T, Obata R (2020) Retinal dystrophy associated with danon disease and pathogenic mechanism through LAMP2-mutated retinal pigment epithelium. Eur J Ophthalmol 30:570–578
Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB (2011) Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity 35:182–193
Gu X, Han M, Du Y, Wu Y, Xu Y, Zhou X, Ye D, Wang HL (2019) Pb disrupts autophagic flux through inhibiting the formation and activity of lysosomes in neural cells. Toxicol Vitro 55:43–50
He JP, Hou PP, Chen QT, Wang WJ, Sun XY, Yang PB, Li YP, Yao LM, Li X, Jiang XD, Chien KY, Zhang ZM, Wu QW, Cowin AJ, Wu Q, Chen HZ (2018) Flightless-I blocks p62-mediated recognition of LC3 to impede selective autophagy and promote breast cancer progression. Cancer Res 78:4853–4864
Hernaez B, Cabezas M, Munoz-Moreno R, Galindo I, Cuesta-Geijo MA, Alonso C (2013) A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med 13:305–316
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222
Kuo ML, Lee MB, Tang M, den Besten W, Hu S, Sweredoski MJ, Hess S, Chou CM, Changou CA, Su M (2016) PYCR1 and PYCR2 interact and collaborate with RRM2B to protect cells from overt oxidative stress. Sci Rep 6:18846
Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A (2009) Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268
Matamoros T, Alejo A, Rodriguez JM, Hernaez B, Guerra M, Fraile-Ramos A, Andres G (2020) African Swine Fever Virus Protein pE199L Mediates Virus Entry by Enabling Membrane Fusion and Core Penetration. MBio 11:e00789-20
McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z (2011) Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem 286:22147–22159
Nakayama T, Al-Maawali A, El-Quessny M, Rajab A, Khalil S, Stoler JM, Tan WH, Nasir R, Schmitz-Abe K, Hill RS (2015) Mutations in PYCR2, encoding Pyrroline-5-carboxylate reductase 2, cause microcephaly and hypomyelination. Am J Hum Genet 96:709–719
Nardacci R, Ciccosanti F, Marsella C, Ippolito G, Piacentini M, Fimia GM (2017) Role of autophagy in HIV infection and pathogenesis. J Intern Med 281:422–432
Ngounou Wetie AG, Sokolowska I, Woods AG, Roy U, Deinhardt K, Darie CC (2014) Protein-protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches. Cell Mol Life Sci 71:205–228
Nguyen HT, Noguchi S, Sugie K, Matsuo Y, Nguyen CTH, Koito H, Shiojima I, Nishino I, Tsukaguchi H (2018) Small-vessel vasculopathy due to aberrant autophagy in LAMP-2 deficiency. Sci Rep 8:3326
Ou R, Zhang X, Cai J, Shao X, Lv M, Qiu W, Xuan X, Liu J, Li Z, Xu Y (2016) Downregulation of pyrroline-5-carboxylate reductase-2 induces the autophagy of melanoma cells via AMPK/mTOR pathway. Tumour Biol 37:6485–6491
Peng H, Liu B, Yves TD, He Y, Wang S, Tang H, Ren H, Zhao P, Qi Z, Qin Z (2018) Zika virus induces autophagy in human umbilical vein endothelial cells. Viruses 10:259
Sun H, Jenson J, Dixon LK, Parkhouse ME (1996) Characterization of the African swine fever virion protein j18L. J Gen Virol 77(Pt 5):941–946
Wang N, Zhao D, Wang J, Zhang Y, Wang M, Gao Y, Li F, Wang J, Bu Z, Rao Z (2019) Architecture of African swine fever virus and implications for viral assembly. Science 366:640–644
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
Yasui LS, Duran M, Andorf C, Kroc T, Owens K, Allen-Durdan K, Schuck A, Grayburn S, Becker R (2016) Autophagic flux in glioblastoma cells. Int J Radiat Biol 92:665–678
Yuan X, Wang B, Yang L, Zhang Y (2018) The role of ROS-induced autophagy in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 42:306–312
Zaki MS, Bhat G, Sultan T, Issa M, Jung HJ, Dikoglu E, Selim L, Imam GM, Abdel-Hamid MS, Abdel-Salam G (2016) PYCR2 Mutations cause a lethal syndrome of microcephaly and failure to thrive. Ann Neurol 80:59–70
Zhang Z, Singh R, Aschner M (2016) Methods for the detection of autophagy in mammalian cells. Curr Protoc Toxicol 69:201221–201226
Zhao D, Liu R, Zhang X, Li F, Wang J, Zhang J, Liu X, Wang L, Zhang J, Wu X, Guan Y, Chen W, Wang X, He X, Bu Z (2019) Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg Microbes Infect 8:438–447
Acknowledgements
The authors thank teacher Wei Shu from the Testing Center of South China Agricultural University for the technical support of laser confocal experiment. This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFD0502303), the Key Research and Development Program of Guangdong Province (2020B020222001), China Postdoctoral Science Foundation (grant no. 2019M652922), the Natural Science Foundation of Guangdong Province (grant no. 2019A1515012006) and the Chief expert Project of Agricultural Industry Technology system in Guangdong Province (2019KJ128).
Author information
Authors and Affiliations
Contributions
SC contributed to the overall experimental design, execution of all laboratory experiments, manuscript writing and interpretation of all data. XZ, HL, WC and WL contributed to experimental design. YN contributed to the plasmid construction and the data analysis. XZ contributed to the manuscript revisions. FC and QX contributed to the financial support and experimental design. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, S., Zhang, X., Nie, Y. et al. African Swine Fever Virus Protein E199L Promotes Cell Autophagy through the Interaction of PYCR2. Virol. Sin. 36, 196–206 (2021). https://doi.org/10.1007/s12250-021-00375-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-021-00375-x