Abstract
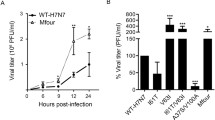
HIV-1 Rev is an accessory protein that plays a key role in nuclear exportation, stabilization, and translation of the viral mRNAs. Rev of HIV-1 clade BC often shows a truncation of 16 AAs due to a premature stop codon at residue 101. This stop codon presents the highest frequency in clade BC and the lowest frequency in clade B. In order to discover the potential biological effect of this truncation on Rev activity and virus replication of clade BC, we constructed Rev expression vectors of clade BC with or without 16 AAs within C-terminal separately, and replaced the stop codon by Q in a CRF07_BC infectious clone. We found that 16 AAs truncation had no effect on expression and activity of Rev in clade BC. Also, the mutation from the stop codon to Q had no effect on virus replication of clade BC. Next, to investigate the effect of this truncation on Rev activity and replication capacity of clade B, Rev expression vectors of clade B carrying or lacking 16 AAs in C-terminal were constructed respectively, and residue Q at position 101 within Rev was substituted by the stop codon in a clade B infectious clone. It was found that 16 AAs truncation significantly down-regulated Rev expression and impaired clade B Rev activity. Furthermore, a Q-to-stop codon substitution within Rev significantly reduced viral replication fitness of clade B. These results indicate that the premature stop codon at residue 101 within Rev exerts diverse impact on viral replication among different HIV-1 clades.






Similar content being viewed by others
References
Booth DS, Cheng Y, Frankel AD (2014) The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Elife 3:e04121
Churchill MJ, Chiavaroli L, Wesselingh SL, Gorry PR (2007) Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus. Retrovirology 4:43
DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, Grimes JM, Wingfield PT, Stuart DI, Steven AC (2016) The structure of HIV-1 rev filaments suggests a bilateral model for Rev-RRE assembly. Structure 24:1068–1080
Dykes C, Wang J, Jin X, Planelles V, An DS, Tallo A, Huang Y, Wu H, Demeter LM (2006) Evaluation of a multiple-cycle, recombinant virus, growth competition assay that uses flow cytometry to measure replication efficiency of human immunodeficiency virus type 1 in cell culture. J Clin Microbiol 44:1930–1943
Fernandes JD, Faust TB, Strauli NB, Smith C, Crosby DC, Nakamura RL, Hernandez RD, Frankel AD (2016) Functional segregation of overlapping genes in HIV. Cell 167:1762–1773
He X, Xing H, Ruan Y, Hong K, Cheng C, Hu Y, Xin R, Wei J, Feng Y, Hsi JH, Takebe Y, Shao Y (2012) A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS ONE 7:e47289
Huang SW, Wang SF, Lin YT, Yen CH, Lee CH, Wong WW, Tsai HC, Yang CJ, Hu BS, Lin YH, Wang CT, Wang JJ, Hu Z, Kuritzkes DR, Chen YH, Chen YM (2014) Patients infected with CRF07_BC have significantly lower viral loads than patients with HIV-1 subtype B: mechanism and impact on disease progression. PLoS ONE 9:e114441
Iversen AK, Shpaer EG, Rodrigo AG, Hirsch MS, Walker BD, Sheppard HW, Merigan TC, Mullins JI (1995) Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol 69:5743–5753
Jackson PE, Tebit DM, Rekosh D, Hammarskjold ML (2016) Rev-RRE functional activity differs substantially among primary HIV-1 isolates. AIDS Res Hum Retrovir 32:923–934
Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY (2005) Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. Cell Res 15:923–934
Ma L, Guo Y, Yuan L, Huang Y, Sun J, Qu S, Yu X, Meng Z, He X, Jiang S, Shao Y (2009) Phenotypic and genotypic characterization of human immunodeficiency virus type 1 CRF07_BC strains circulating in the Xinjiang Province of China. Retrovirology 6:45
Ma J, Dykes C, Wu T, Huang Y, Demeter L, Wu H (2010) vFitness: a web-based computing tool for improving estimation of in vitro HIV-1 fitness experiments. BMC Bioinform 11:261
Malim MH, Bohnlein S, Hauber J, Cullen BR (1989) Functional dissection of the HIV-1 Rev trans-activator–derivation of a trans-dominant repressor of Rev function. Cell 58:205–214
Phuphuakrat A, Auewarakul P (2005) Functional variability of Rev response element in HIV-1 primary isolates. Virus Gen 30:23–29
Phuphuakrat A, Paris RM, Nittayaphan S, Louisirirotchanakul S, Auewarakul P (2005) Functional variation of HIV-1 Rev response element in a longitudinally studied cohort. J Med Virol 75:367–373
Pollard VW, Malim MH (1998) The HIV-1 Rev protein. Annu Rev Microbiol 52:491–532
Rausch JW, Le Grice SF (2015) HIV Rev assembly on the Rev response element (RRE): a structural perspective. Viruses 7:3053–3075
Smith AJ, Cho MI, Hammarskjold ML, Rekosh D (1990) Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol 64:2743–2750
Su L, Graf M, Zhang Y, von Briesen H, Xing H, Kostler J, Melzl H, Wolf H, Shao Y, Wagner R (2000) Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B’) recombinant strain in China. J Virol 74:11367–11376
Toth-Petroczy A, Palmedo P, Ingraham J, Hopf TA, Berger B, Sander C, Marks DS (2016) Structured states of disordered proteins from genomic sequences. Cell 167(158–170):e112
Wang J, Bambara RA, Demeter LM, Dykes C (2010) Reduced fitness in cell culture of HIV-1 with nonnucleoside reverse transcriptase inhibitor-resistant mutations correlates with relative levels of reverse transcriptase content and RNase H activity in virions. J Virol 84:9377–9389
Wang Z, Hong K, Zhang J, Zhang L, Li D, Ren L, Liang H, Shao Y (2013) Construction and characterization of highly infectious full-length molecular clones of a HIV-1 CRF07_BC isolate from Xinjiang, China. PLoS ONE 8:e79177
Wang Z, Zhang J, Li F, Ji X, Liao L, Ma L, Xing H, Feng Y, Li D, Shao Y (2017) Drug resistance-related mutations T369 V/I in the connection subdomain of HIV-1 reverse transcriptase severely impair viral fitness. Virus Res 233:8–16
Wu Y, Wang H, Ren X, Wan Z, Hu G, Tang S (2017) HIV-1 CRF07_BC with a seven amino acid deletion in the gag p6 region dominates in HIV-1-infected men who have sex with men in China. AIDS Res Hum Retrovir 33:977–983
Yang R, Kusagawa S, Zhang C, Xia X, Ben K, Takebe Y (2003) Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J Virol 77:685–695
Zhefeng M, Huiliang H, Chao Q, Jun S, Jianxin L, Xiaoyan Z, Jianqing X (2011) Transmission of new CRF07_BC strains with 7 amino acid deletion in Gag p6. Virol J 8:60
Acknowledgements
We appreciate the generous gifts of AT1 and AT2 plasmids from Dr. Carrie Dykes (The University of Rochester School of Medicine and Dentistry), as well as BH10-Rev expression plasmid and pCMV-Gag-Pol-RRE expression plasmid of clade B from Dr. Min Wei (Nankai University). Clade BC Gag-Pol-RRE expression vector was received from Dr. Jingwan Han (Beijing Institute of Microbiology and Epidemiology). CMV-EGFP plasmid was gifted from Dr. Lei Yu (Shenzhen Second People’s Hospital). We would like to thank Hong Peng (The Division of Research of Virology and Immunology, National Center for AIDS/STD Control and Prevention) for the technical supports for this work. This study was partially funded by the National Natural Science Foundation of China (Grants 81872680 and 31600734), the Yong Scientific Research Foundation of NCAIDS/STD (Grant 2018AFQN002) and the SKID outstanding youth grant (2019SKLID402). Funding has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
ZW and YS conceived and designed the experiments. ZW and XJ performed the experiments. ZW, XJ and KH analyzed the data. DL, YH and YS contributed reagents/materials/analysis tools. ZW, KH and LM wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
Informed consent have been obtained from all participants and the studies have been approved by the Institutional Review Board of the National Center for AIDS/STD control and Prevention, China CDC.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Z., Ji, X., Hao, Y. et al. Premature Stop Codon at Residue 101 within HIV-1 Rev Does Not Influence Viral Replication of Clade BC but Severely Reduces Viral Fitness of Clade B. Virol. Sin. 35, 181–190 (2020). https://doi.org/10.1007/s12250-019-00179-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-019-00179-0