Abstract
Porcine deltacoronavirus (PDCoV) is a newly identified virus that causes watery diarrhea in newborn piglets and results in significant economic losses to the pig industry. Since first reported in Hong Kong in 2012, PDCoV has been subsequently detected in USA, South Korea, Thailand, and mainland China. Here we isolated a strain of PDCoV, named CHN-GD-2016, from the intestinal content of a diseased newborn piglet with severe diarrhea in a pig farm in Guangdong, China. PDCoV CHN-GD-2016 could be identified by immunofluorescence with PDCoV specific rabbit antisera, and typical crown-shaped particles with spiky surface projections of this PDCoV were observed with electron microscopy. Genomic analysis showed that the PDCoV CHN-GD-2016 was closely related to other Chinese PDCoV strains, with the highest sequence similarity with the strain CHN/Tianjin/2016. Importantly, inoculation of newborn piglets with 1 × 105 TCID50 of CHN-GD-2016 by oral feeding successfully reproduced clear clinical symptoms, including vomiting, dehydration, and severe diarrhea in piglets. In addition, the virus RNA in rectal swabs from 1 to 7 days post inoculation was detected, macroscopic and microscopic lesions in small intestine were observed, and viral antigen was also detected in the small intestines with immunohistochemical staining. Collectively, the data show in this study confirms that PDCoV is present in Guangdong, China and is highly pathogenic in newborn piglets.
Similar content being viewed by others
Introduction
Coronaviruses (CoVs) are found in a wide variety of animals. They belong to the subfamily Coronavirinae in the family Coronaviridae within the order Nidovirales (Zhang 2016). Porcine CoVs are significant enteric and respiratory pathogens of swine. Five porcine CoVs have so far been identified: transmissible gastroenteritis virus (TGEV) (Doyle and Hutchings 1946), porcine respiratory coronavirus (PRCV) (Wesley et al. 1990), and porcine epidemic diarrhea virus (PEDV) (Pensaert and De Bouck 1978) in the Alphacoronavirus genus; porcine hemagglutinating encephalomyelitis virus (PHEV) (Sasseville et al. 2002) in the Betacoronavirus genus; and porcine deltacoronavirus (PDCoV) (Woo et al. 2012) in the Deltacoronavirus genus. PDCoV was first detected in 2012 in Hong Kong during a molecular epidemiology study of CoVs using RT-PCR to examine the pig samples and complete genome sequences of these two PDCoV strains (HKU15-44, HKU15-155) were reported (Woo et al. 2012). PDCoV is an enveloped, single-stranded, positive-sense RNA virus with a genome of appropriately 25 kb in length (Zhang 2016). The genome organization of PDCoV are in the order of: 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), nonstructural protein 7 (NS7), and 3′ UTR (Chen et al. 2015a, b). The clinical symptoms in newborn piglets caused by PDCoV infection are similar to that by other porcine enteric pathogens such as PEDV and TGEV, which includes vomiting, diarrhea, dehydration and death (Zhang 2016). To date, there are no effective treatments or vaccines for PDCoV.
Since first detection in pig feces in 2012 (Woo et al. 2012), PDCoV subsequently has been reported in at least nine states of the United States, Thailand, Vietnam, Lao PDR, South Korea, and Canada (Lee and Lee 2014; Marthaler et al. 2014; Wang et al. 2014a, b; Saeng-Chuto et al. 2017). Apart from these places, PDCoV was also detected in mainland China (Chen et al. 2015a, b; Song et al. 2015; Dong et al. 2016; Wang et al. 2015). These molecular surveillance studies indicated that PDCoV was a common viral pathogen of pigs around the world. Increasing virological and serological methods have been developed to detect PDCoV in clinical samples from pig farms. Virological methods include detection of virus particles by electron microscopy (Ma et al. 2015), detection of viral nucleic acid by RT-PCR and real-time PCR (Marthaler et al. 2014; Hu et al. 2015a, b). And the most commonly used serological assays for PDCoV are indirect fluorescent antibody (IFA) assay (Dong et al. 2016) and enzyme linked immunosorbent assay (ELISA) targeting PDCoV nucleocapsid (N) protein (Su et al. 2016).
Currently several studies have reproduced clinical diarrheal disease by experimentally infecting gnotobiotic and conventional pigs with isolated PDCoV strains (Chen et al. 2015a, b; Jung et al. 2015; Ma et al. 2015; Dong et al. 2016). As expected, the 11–14 day-old gnotobiotic pigs inoculated orally with PDCoV strain OH-FD22 and OH-FD100 showed severe vomiting, diarrhea, and atrophic enteritis (Jung et al. 2015). All infected pigs had PEDV-like lesions characterized by thin and transparent intestinal walls and accumulation of large amounts of yellow fluid in the intestinal lumen (Jung et al. 2015). Further, histology analysis exhibited acute diffuse, severe atrophic enteritis and mild vacuolation of superficial epithelial cells in cecum and colon (Jung et al. 2015). The pathogenicity of PDCoV in conventional pigs by subsequent isolation was also confirmed (Chen et al. 2015a, b; Ma et al. 2015; Dong et al. 2016).
Although PDCoV was detected in southern China (Zhai et al. 2016), detailed information of PDCoV isolates from southern China remains unclear. In this study, we isolated a PDCoV strain from Guangdong province of China using LLC-PK cells, characterized its complete genome by sequencing, and investigated its pathogenicity in 5-day-old conventional pigs by clinical assessment, the quantification of viral shedding, histological and immunohistochemical assays. The results suggested that the isolate of PDCoV CHN-GD-2016 was closely related to other PDCoV strains from mainland China and was capable of causing severe watery diarrhea in piglets.
Materials and Methods
Clinical Samples Collection and Specific RT-PCR Testing
From June to October 2016, 24 intestinal contents were collected from newborn piglets on different seven commercial pig farms with reported diarrhea outbreaks in Guangdong, China, and stored at −80 °C until further use. Prior to viral RNA extraction, intestinal contents were diluted one time using sterile 1 × phosphate buffer saline (PBS) (pH 7.4). The supernatants were then collected by centrifugation at 6010 ×g for 5 min at 4 °C. Total RNA was prepared from the supernatants using a RNeasy kit (Magen, China) according to the manufacturer’ instruction, and was treated with DNase I. The specific primers for the nucleocapsid (N) gene of PDCoV (sense: 5′-ATGGCTGCACCAGTAGTCCCTA-3′; antisense: 5′-GGAGCTTGATGCTGGGTCTTG-3′) were designed with reference to the published sequence (GenBank, Accession no: JQ065043), the nucleocapsid (N) gene of PEDV (sense: 5′-TTTCTAAGGTACTTGCAAATAATG-3′; antisense: 5′-TTGGAGATCTGGACCTGTTGTTGC-3′), the nucleocapsid (N) gene of TGEV (sense: 5′-TCATGCAGATGCCAAATTTAAAGA-3′; antisense: 5′-TCATCCTTCTTGTTATTGAATTGT-3′), and the vp6 gene of PRoV (sense: 5′-CTCGATGCTACTACAGAATCAG-3′; antisense: 5′-AGCCACATAGTTCACATTTCATCC-3′) were designed with reference to previous publications (Jeong et al. 2009; Hu et al. 2015a, b) and synthesized by Sangon Company (Shanghai, China). The one step RT-PCR was performed in a 25-μL volume containing 2 μg of RNA, 1 μL of PrimeScript 1 Step Enzyme Mix, 12.5 μL 2 × 1 Step Buffer (TaKaRa, Dalian), and a 0.4 μmol/L of each gene-specific primer. The thermal cycling parameters were as follows: 50 °C for 30 s, 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 10 min and all the positive constructs were confirmed with sequencing analysis by Sangon Company (Shanghai, China). The RNA of PRRSV (PRRSV strain was kindly provided by Professor Yaosheng Chen (Sun Yat-sen University)) or PEDV (PEDV GDS01 strain was isolated in our lab) as controls.
Virus Isolation, Plaque Purification and Propagation in LLC-PK Cells
LLC-PK cells were obtained from Guangdong Wen’s Foodstuffs Group Co., Ltd (Guangdong, China) and were used to isolate PDCoV from the original positive samples. LLC-PK cells were cultured in DMEM (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (BOVOGEN, Australia), and the maintenance medium for PDCoV propagation was DMEM supplemented with 8 μg/mL trypsin (Gibco, USA) in 5% CO2 incubator.
Virus isolation, plaque purification and propagation were performed as previously described with some modifications (Hu et al. 2015a, b). Briefly, for the first inoculation, LLC-PK cells were cultured in 6-well plates, and were washed three times with DMEM when 90% confluent of cell monolayers. Two hundred microliter of the filtered inoculums, together with 350 μL maintenance medium was added to each well. After adsorption for 1.5 h at 37 °C in 5% CO2, cells were washed 3 times with maintenance medium, and 2 mL maintenance medium was added. The cells were cultured continuously at 37 °C in 5% CO2 to observe cytopathic effect (CPE). When > 80% CPE was evident in the inoculated cell monolayers (around day 6 postinoculation (p.i.) after the first inoculation), the plates were frozen at −80 °C and thawed twice. The cells and supernatants were harvested together, the 0-h postinoculation and day 6 p.i. samples were tested by PDCoV specific RT-PCR as above described. These samples were used as seed stocks for the next passage and plaque purification. For virus plaque purification, supernatants from virus-infected cells were serially diluted and used to infect LLC-PK cells in maintenance medium for 1.5 h at 37 °C in 5% CO2 and then the maintenance were discarded, followed by overlaying 2 mL maintenance medium containing 1.25% Agarose LM GQT (TaKaRa, Dalian) to immobilize the virus. After 24 h, cells were fixed and visualized with 2 mL maintenance medium containing 1.25% Agarose LM GQT and 0.01% Neutral red solution (Sigma, USA), and the plaques were picked by using sterile pipette tips, and the agarose plug was placed into a microcentrifuge tube containing 0.5 mL maintenance medium. The selected plaques of PDCoV were named CHN-GD-2016 and used for viral propagation. LLC-PK cells were cultured in T175 flasks, and they were washed three times with maintenance medium when 90% confluent cell monolayers. Fifty microliters of PDCoV together with 50 mL maintenance medium were added into flask. The cell pellets and supernatants were cultured continuously at 37 °C in 5% CO2 to observe CPE. When CPE was evident in the inoculated cell monolayers (around day 1 p.i.), the plates were frozen at −80 °C and thawed twice. The cells and supernatants were harvested together to determine viral titers.
Infectious-Virus Titrations by a TCID50 Assay
LLC-PK cells were seed on 96-well plates and cultured overnight before washed two times with the maintenance medium. One hundred microliter of tenfold dilutions of PDCoV was inoculated in eight replicates per dilution, then the cells were cultured continuously at 37 °C in 5% CO2. Viral CPE was observed for 5–7 days, and virus titer were calculated by using the Reed–Muench method (Reed and Muench 1938) and expressed as TCID50 per milliliter.
Immunofluorescence Assay (IFA)
Immunofluorescence assay was conducted to observe PDCoV-infected LLC-PK cells as described previously with some modifications (Dong et al. 2016). Briefly, LLC-PK cells (1 × 105) were seeded on 24-well plates and were cultured overnight before mock infected or infected with plaque-purified PDCoV at a multiplicity of infection (MOI) of 0.01. At 24 h after inoculation, the cells were fixed with 4% paraformaldehyde for 20 min and then permeabilized with 0.2% Triton X-100 for 10 min at room temperature. The cells were then blocked with 1% bovine serum albumin (BSA), and incubated with PDCoV specific rabbit antisera (Guangdong Wen’s Foodstuffs Group Co., Ltd, China) (1:100), followed by fluoresceinisothiocyanate (FITC)-labeled goat anti-rabbit secondary antibody (KPL, USA) (1:1000) for 1 h. The cell nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) [Beyotime Biotechnology (Nanjing, China)] for 3 min at room temperature. After the cells were washed with 1 × PBS, the stained cells were observed with a fluorescence microscope (LEICA TCS SP5, Germany).
Electron Microscopic Observation
Electron microscopy (EM) was conducted to observe virus samples as described previously with some modifications (Kong et al. 2013; Hu et al. 2015a, b). For visualization of the virion particles in infected-cell culture medium, PDCoV-infected LLC-PK cells were frozen at −80 °C and thawed twice, and the cell culture clarified by centrifugation at 7012 ×g for 30 min at 4 °C. The supernatant were supplemented with 6% PEG6000 12 h at 4 °C. The mixture were centrifuged at 11,180 ×g for 1 h at 4 °C, and the pellet was resuspended in sterile 1 × PBS (pH 7.4) buffer, followed to equilibrium in 8 mL non-linear 20%–60% sucrose-TNE gradients by centrifugation at 106,750 ×g for 2 h at 4 °C by using an ultracentrifuge (Hitachi Koki himac CP 100WX, Japan). After purifition with sucrose gradient centrifugation, purified virions were diluted with sterile 1 × PBS (pH 7.4) buffer and removed the sucrose by centrifugation at 7012 ×g for 2 h at 4 °C by using centrifugal filter units (Millipore, USA). The purified virus pellets were resuspended in sterile 1 × PBS (pH 7.4) buffer and negatively stained with 3% phosphotungstic acid. After blotting and drying, the grids were examined with a JEM-100 CX-II electron microscope (JEOLLTD, Japan).
Complete Genomic Analysis
After we successfully isolated the PDCoV strain CHN-GD-2016, the complete genome was sequenced by Illumina HiSeq as previously described with some modifications (Malboeuf et al. 2013). Briefly, total RNA was prepared from the PDCoV-infected LLC-PK cells using a RNeasy kit (Magen, China) per the manufacturer’ instruction, and was purified as described above. cDNA synthesis was performed for sequencing library preparation by reverse transcription using RT-PCR kit (TaKaRa, Dalian). The genomic cDNA was disrupted and converted to a sticky end by added the A-base at 3′ end of the cDNA. The DNA containing Index sequence was added at either side of sticky end by complementary bases. The target fragment within the scope of a certain length was collected before magnetic beads selection. The sequencing of library was builded and tested using PCR amplification Index sequence, and the sequencing of library combined to the chip by bridge PCR. At last, the complete genome was sequenced by IIIumina HiSeq. Sequence alignments spike genes of different porcine deltacoronavirus (PDCoV) strains were performed using the DNAStar Lasergene 7.0. A genome homology analysis and phylogenetic trees were constructed by using the maximum likelihood method with MEGA 5 software (http://www.megasoftware.net/) based on the whole-genome nucleotide sequences of 27 PDCoV strains from different countries.
Experimental Infection with the PDCoV CHN-GD-2016 Strain in Conventional Newborn Piglets
The animal study was approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University and used in accordance with regulation and guidelines of this committee. Eight 4-day-old conventional newborn piglets were purchased from a conventional commercial pig farm and randomly divided into two groups (4 piglets/group) and were housed in two separate rooms. Piglets were fed a mixture of skim milk powder (Inner Mongolia Yili Industrial Group Co., Ltd, China) with warm water. Prior to inoculation, piglets were confirmed negative for the major procine enteric viruses (PDCoV, PEDV, TGEV, PRoV) by testing of rectal swabs using specific RT-PCR as above described. After 1 day acclimation, piglets in group 1 were orally inoculated with 10 mL of maintenance medium and served as uninfected controls. Piglets in group 2 were orally challenged with 10 mL of maintenance medium containing 1 × 105 TCID50 of the PDCoV CHN-GD-2016 strain. Piglets were observed daily for clinical signs of vomiting, diarrhea, lethargy, body weight. Rectal swabs were collected daily from each piglet from day 1 p.i. to necropsy and were submerged into 1 mL PBS immediately after collection. All piglets from each group were necropsied at day 7 p.i.. At necropsy, the fresh heart, lung, spleen, liver, kidney, jejunum were collected and fixed by 10% formalin for histopathology and immunohischemistry analysis.
Real-Time RT-PCR Analysis
The supernatants of rectal swab from each piglet centrifuged at 6010 ×g for 5 min. Total RNA was prepared from the supernatants using a RNeasy kit (Magen, China) per the manufacturer’ instruction, and was treated with DNase I. Two μg of total RNA was used for cDNA synthesis by reverse transcription using RT-PCR kit (TaKaRa, Dalian). The specific primers for the membrane (M) gene of PDCoV (sense: 5′-ATCGACCACATGGCTCCAA-3′; antisense: 5′-CAGCTCTTGCCCATGTAGCTT-3′), and probe (5′-FAM-CACACCAGTCGTTAAGCATGGCAAGCT-BHQ-3′) were designed with reference to the previous publication (Marthaler et al. 2014) and synthesized by Invitrogen Company (Shanghai, China). The real-time PCR assay was carried out with an Applied Biosystem 7500 Fast instrument (Life Technologies, USA). The PCR was performed in a 20-μL volume containing 1 μL of cDNA, 10 μL of Thunderbird Probe qPCR Mix, 0.04 μL 50 × Rox reference dye (TOYOBO, Shanghai), 0.2 μmol/L of probe, and a 0.3 μmol/L of each gene-specific primer. The thermal cycling parameters were as follows: 95 °C for 20 s; 40 cycles of 95 °C for 3 s, 60 °C for 30 s. The standard curve generated by construction of plasmids. Briefly, the M gene was amplified from PDCoV CHN-GD-2016 strain using the specific primers (sense: 5′-ATGTCTGACGCAGAAGAGTG-3′; antisense: 5′-TTACATATACTTATACAGGCGAGC-3′) that were designed with reference to the published sequence (GenBank, Accession no: JQ065043), and the PCR products were cloned into the pMD19-T (TaKaRa, Dalian). The known plasmid concentration was tenfold serially diluted for generating a standard curve in each plate. The quantity of PDCoV viral RNA in tested samples was calculated based on the cycle threshold (Ct) values for the standard curve.
Histological and Immunohistochemical Staining
At necropsy, tissue samples of heart, lung, spleen, liver, kidney and jejunum of the piglets from the challenged and control groups were separated and routinely fixed in 10% formalin for 36 h at room temperature (Hu et al. 2016), and then dehydrated in graded ethanol, embedded in paraffin, cut in 5-μm sectioned, and mounted onto glass slides. After the sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E), the slides were examined and analyzed with conventional light microscopy. Section (5 μm) of formalin-fixed paraffin-embedded tissues were placed onto positively charged glass slides and the slides were air dried for 120 min at 60 °C. The tissue sections were deparaffinized, and then rinsed and incubated with target retrieval solution (Servicebio, China). After the sections were blocked with 1% BSA (Solarbio, China), they were incubated with PDCoV specific rabbit antisera (Guangdong Wen’s Foodstuffs Group Co., Ltd, China) (1:100) as the primary antibody for 12 h at 4 °C. They were then incubated with peroxidase-labeled goat anti-rabbit IgG secondary antibody (KPL, USA) (1:200) for 50 min at room temperature, and the samples were finally visualized with a 3, 3′-diaminobenzidine (DAB) chromogen kit (Dako, Denmark). Hematoxylin was used for counterstaining. Tissues of piglets from negative control groups were used as negative samples.
Results
Prevalence of PDCoV in Guangdong, China
Diarrhea occurred in the piglets on conventional pig farms in southern China from 2012 to 2015 (Zhai et al. 2016). To determine the causative agent of the diarrhea outbreaks in Guangdong province of China, 24 clinical samples were collected from newborn piglets on seven different commercial pig farms with reported diarrhea outbreaks from June to October 2016, and used to detect the presence of porcine viral enteric pathogens (PDCoV, PEDV, TGEV and Rota C). Of 24 intestinal contents examined, 4 (16.7%) were PDCoV positive (Fig. 1A), 11 (45.8%) were PEDV positive (Fig. 1B), and 2 (8.3%) were positive for both PDCoV and PEDV, indicating that PDCoV/PEDV coinfections exist in pig farms. Interestingly, none of these samples were positive for TGEV or PRoV (data not shown), indicating that PDCoV or PEDV is the main cause of diarrhea in piglets in Guangdong, China. These data suggest that PDCoV was present in Guangdong province of China despite a relatively lower prevalence.
A Strain of PDCoV was Isolated from a Diarrheic Newborn Piglet
Since the samples were examined as PDCoV-positive, we attempted to isolate PDCoV from the positive samples. LLC-PK cells monolayers were inoculated with 2 of the samples that were positive for PDCoV only according to the published method (Hu et al. 2015a, b). Compared with the control in Fig. 2A, one inoculated cell monolayers showed visible CPE in the form of rounded and clustered cells at day 6 p.i. (Fig. 2B). To confirm PDCoV replication in LLC-PK cells, viral RNA was extracted from the inoculated cells at day 6 p.i. and tested by specific RT-PCR. This cell culture-passaged sample was positive for PDCoV but negative for other porcine enteric viruses (data not shown). PDCoV from the first passage in LLC-PK cells was named CHN-GD-2016. The PDCoV CHN-GD-2016 was further serially passaged in LLC-PK cells for a total of 10 passages, and was still confirmed PDCoV positive by specific RT-PCR (data not shown), indicating that PDCoV CHN-GD-2016 strain could replicate in LLC-PK cells.
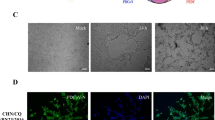
Cytopathic effects (CPE) and IFA staining on PDCoV-inoculated LLC-PK cells. A Mock-inoculated LLC-PK cell culture showing normal cells. B PDCoV inoculated LLC-PK cells at day 6 p.i. showing rounded and clustered cells (indicated by arrows). LLC-PK cells were mock-inoculated (C) or inoculated with PDCoV CHN-GD-2016 (D). At 24 h postinoculation, an immunofluorescence assay (IFA) was performed (PDCoV antigen was indicated by arrows).
To determine this result, the plaque-purified PDCoV in LLC-PK cells was further confirmed by IFA with PDCoV specific rabbit antisera. As shown in Fig. 2C–D, PDCoV-specific immunofluorescence was detected. Taken together, these results clearly show that we successfully isolated a PDCoV strain from the sample. To characterize the morphology and size of virus particles, the PDCoV CHN-GD-2016 virus purified from infected LLC-PK cells were examined with electron microscopy (EM). Typical crown-shaped particles with spiky surface projections of coronavirus were observed by negative staining on EM, being 80–160 nm in diameter (Fig. 3). Taken together, these results clearly showed that a PDCoV strain was successfully isolated from the intestinal contents of a newborn diarrheic piglet in Guangdong, China.
Complete Genome Sequencing and Phylogenetic Analysis of PDCoV CHN-GD-2016
Successful isolation of CHN-GD-2016 prompted us to characterize its complete genome (GenBank no: MF280390). The complete genome showed that PDCoV strain CHN-GD-2016 share 97.3%–99.5% nucleotide identity with the other 26 PDCoV strains in GenBank. Compared with those PDCoV strains, a 3-nt deletion was observed in the S gene of PDCoV strain CHN-GD-2016, which was also present in all PDCoV strains from mainland China except CHN-AH-2004 (Fig. 4A). To further analyze the PDCoV strain CHN-GD-2016, a phylogenetic tree of 27 PDCoV strains was constructed using the neighbor-joining method in the MEGA 5 program. Phylogenetic analysis showed that the PDCoV strains from the United States and South Korea were clustered into a large clade, whereas PDCoV strain CHN-GD-2016 clustered with other PDCoV strains detected in China since 2014 (Fig. 4B). Therefore, these data suggested that the CHN-GD-2016 strain was most closely related to other PDCoV strains from mainland China.
Sequence alignments partial spike genes of different porcine deltacoronavirus strains and phylogenetic tree constructed on the basis of the whole-genome nucleotide sequences of 27 PDCoV strains from different countries. A A multiple sequence of partial spike (S) genes was constructed with Clustal W in the DNAStar software. Red rectangles indicate the 3-nt deletion sites. B The dendrogram was constructed using the neighbor-joining method in the MEGA software package, version 5 (http://www.megasoftware.net). Bootstrap resampling (1000 replication) was performed, and bootstrap values are indicated for each node. Reference sequence obtained from GenBank are indicated by strain name. The scale bar represents 0.002 nucleotide substitutions per site.
PDCoV CHN-GD-2016 Strain was Highly Pathogenic to Newborn Piglets
In order to determine whether the newly-isolated strain of PDCoV CHN-GD-2016 was a causative agent for diarrhea, we experimentally infected newborn piglets with the isolated virus to reproduce diarrhea. As expected, the newborn piglets inoculated with CHN-GD-2016 at a dose of 1 × 105 TCID50/head via oral feeding showed typical clinical syndrome, characterized by acute and severe watery diarrhea, vomiting, and mild dehydration, as compared with controls (Fig. 5A, B), indicating that the isolated PDCoV CHN-GD-2016 strain might serve as an important cause of severe watery diarrhea in newborn piglets. Importantly, the PDCoV RNA was detected by qRT-PCR in fecal swabs collected from orally inoculated piglets on day 1 p.i. to day 7 p.i., and no PDCoV RNA was detected in the negative control piglets during the study (Fig. 5C). Taken together, these results demonstrated that PDCoV CHN-GD-2016 strain was highly pathogenic to the newborn piglets.
Reproduction of watery diarrhea and fecal viral shedding in newborn piglets inoculated with PDCoV CHN-GD-2016 strain via oral feeding. A Four newborn piglets uninfected as control. B Watery diarrhea (indicated by arrows) was observed at day 3 p.i. with PDCoV infection. C Ct values of group PDCoV inoculation newborn piglet fecal swabs and viral RNA shedding in fecal swabs after PDCoV inoculation or mock inoculation.
Gross Pathology, Histopathology, and Immunohistochemistry in Newborn Piglets Infected with PDCoV CHN-GD-2016
To determine the gross pathological and histological changes in piglets infected with the PDCoV CHN-GD-2016 strain, all piglets were necropsied at day 7 p.i.. Gross findings were similar in all four piglets orally inoculated with PDCoV CHN-GD-2016. The small intestines, where yellow watery contents had accumulated were transparent, thin-walled, and gas-distended (Fig. 6A). No lesions were observed in any other organs of the PDCoV-challenged piglets or in the organs in the negative control piglets (Fig. 6B), indicating that small intestines is the target organ of PDCoV infection. Microscopic lesions were also analysed. As shown in Fig. 6C, blunt intestinal villus and intestinal lamina propria hyperemia was observed, but the intestinal in negative control was normal (Fig. 6D). Consistent with the histopathological results, the PDCoV antigen in the cytoplasm of the villous enterocytes of the PDCoV-challenged piglets was detected by immunohistochemical analysis (Fig. 6E), but no PDCoV antigen was detected in negative control (Fig. 6F). Taken together, these results indicate that PDCoV CHN-GD-2016 could cause intestinal lesion in newborn piglets.
Intestinal changes in newborn piglets inoculated with PDCoV strain CHN-GD-2016. A Thin-walled small intestines (indicated by arrows) of a PDCoV-challenged newborn piglets at day 7 p.i.. B Macroscopic picture of a control piglet at day 7 p.i.. C Hematoxylin and eosin (H&E)-stained jejunum tissue section of a PDCoV-challenged piglet at day 7 p.i. (Blunt intestinal villus and hyperemia were indicated by arrows). D H&E-stained jejunum tissue section of a control piglet at day 7 p.i.. E Immunohistochemically stained jejunum tissue section of a PDCoV-challenged piglet at day 7 p.i. (PDCoV antigen was indicated by arrows). F Immunohistochemically stained jejunum tissue section of a control piglet at day 7 p.i.
Discussion
Since the first report of PDCoV in pigs in 2012 in Hong Kong (Woo et al. 2012), this novel swine enteric coronavirus has been widely detected and isolated in pig farms in different countries (Lee and Lee 2014; Marthaler et al. 2014; Wang et al. 2014a, b, 2015; Saeng-Chuto et al. 2017). The prevalence of PDCoV in pigs in mainland China was reported to be 23.4% in three provinces in China (Chen et al. 2015a, b), and Song et al. reported a PDCoV positive rate of 33.71% in Jiangxi province, China (Song et al. 2015). Our result showed a PDCoV positive rate of 16.7% in 7 diarrhea-outbreak pig farms in Guangdong, China. In addition, there were higher detection rates of PEDV (45.8%) than of TGEV (0%) or PRoV (0%) in those pig farms. Notably, all two PDCoV-positive piglets were co-infected with PEDV. Although the prevalence of PDCoV in southern China was reported (Zhai et al. 2016), there are no published papers reporting the successful isolation of PDCoV in Guangdong, China. In the present study, we reported that an isolate from a case of piglet diarrhea in Guangdong, was highly pathogenic to newborn piglets. This PDCoV strain can be further used for virological and serological assay development, and vaccine development.
LLC-PK, a stable epithelial-like pig kidney cell line (Hull et al. 1976), is commonly used to isolate PDCoV (Hu et al. 2015a. b; Dong et al. 2016). In addition, ST cell line is also used to isolate PDCoV (Hu et al. 2015a, b). Since the intestinal content samples were confirmed as PDCoV-positive, we attempted to isolate virus from these samples in LLC-PK cells. However, only the PDCoV CHN-GD-2016 strain was successfully isolated in LLC-PK cells. The difficulty to isolate PDCoV from positive samples might be associated with the fact that positive RT-PCR samples may contain noninfectious or low amount of virus (Hu et al. 2015a, b). And the amount of trypsin in LLC-PK cells might also contribute to successful PDCoV isolation. CPE was firstly observed in inoculated LLC-PK cells at day 6 p.i.. After plaque purification and several passages, the viral titer reached 1 × 109 TCID50/mL, showing that the PDCoV CHN-GD-2016 strain was highly replicative in LLC-PK cells. The plaque-purified PDCoV strain in LLC-PK cells was further confirmed by IFA with PDCoV specific rabbit antisera, indicating that we successfully isolated a PDCoV strain in Guangdong. We also observed characteristic crown-like particles of the purified PDCoV CHN-GD-2016 strain by electron microscopy. Despite the successful isolation of one strain, the success rate was relatively low. As a result, further attempts are needed to improve the situation. The attempts might include inoculation of sensitive piglets by fresh homogenate to increase the virus titers for isolation, which are proved to be feasible in PEDV isolation (Chen et al. 2016).
To characterize the virus isolate, the complete genome of the PDCoV CHN-GD-2016 strain was sequenced and analyzed. Based on the phylogenetic tree analysis, all PDCoV strains from GenBank share high nucleotide identities. Notably, the PDCoV CHN-GD-2016 strain can be clustered into one clade with other isolates from mainland China, indicating that PDCoV strains currently circulating in multiple Chinese provinces are closely related. The PDCoV strains isolated in mainland China belong to an independent subcluster. Obviously, further studies are needed to identify the exact origin of the mainland China PDCoVs. Previous studies have shown that the S gene, the most variable region in the coronavirus genomes, belongs to type I membrane glycoproteins family (Woo et al. 2010) and that the S protein of CoVs are involved in receptor binding and viral entry (Woo et al. 2010). Sequencing of S genes revealed characteristic 3 nucleotide deletion, which is also present in most PDCoV strains from mainland China like CHN-HN-2014 strain (Dong et al. 2016), compared with the HKU15 strains. However, whether these unique variations contribute to the efficiency of viral replication and virulence needs to be further investigated.
Of note, several groups have confirmed the pathogenicity of PDCoV strains in gnotobiotic or conventional pigs (Chen et al. 2015a, b; Jung et al. 2015; Ma et al. 2015; Dong et al. 2016). Collectively, these results confirmed that PDCoV cause severe diarrhea and vomiting in pigs of 5–21 days old. We hypothesized that the PDCoV CHN-GD-2016 strain was also capable of causing the same clinical symptoms in piglets. We infected the 5-day-old newborn piglets with the PDCoV CHN-GD-2016 strain via oral feeding. Our success in reproduction of severe diarrhea and vomiting in piglets by oral infection strongly suggested that PDCoV posed a huge threat to the newborn piglets in pig farms. Furthermore, we collected fecal swabs from the PDCoV-challenged piglets and detected the viral fecal shedding with real-time PCR. PDCoV RNA was detected from day 1 p.i. to day 7 p.i., while no RNA was detected in the negative control piglets during the study, indicating that the infection with PDCoV by fecal-oral contamination might be the major mode of transmission for diarrhea in piglets in pig farms. In addition, gross lesions were obviously observed in the small intestines of the 5-day-old piglets at necropsy at day 7 p.i., similar to observations in other studies (Chen et al. 2015a, b; Jung et al. 2015; Ma et al. 2015; Dong et al. 2016). While microscopic lesions were observed in all sections of small intestine infected by PEDV in previous report (Madson et al. 2014), we only observed microscopic lesions in the jejunum and ileum in CHN-GD-2016 infected piglets, indicating that disease caused by PDCoV was milder than that caused by PEDV. In addition, the PDCoV antigen in the cytoplasm of the villous enterocytes of the PDCoV-challenged piglets was detected by immunohistochemical analysis. Together, all these results confirmed that the PDCoV CHN-GD-2016 strain isolated in this study could cause enteric diseases in newborn piglets. However, there are still several important questions needed to be addressed. For example, what is the relationship between this virus and PEDV, the causative agent for severe diarrhea in pigs? And what is the role of the nucleotide changes of S gene on viral replication and virulence? How to prepare effective vaccine against the PDCoV? Elucidation of these questions will elevate our understandings of the pathogenicity of PDCoV infection and may help to develop better strategies to control PDCoV.
In summary, we isolated a field strain of PDCoV from the intestinal content of a severe diarrheic piglet in Guangdong, China. Furthermore, genomic analysis showed that the isolate was closely related to other Chinese PDCoV strains, with the highest sequence similarity with the strain CHN/Tianjin/2016. Importantly, inoculation of newborn piglets with PDCoV CHN-GD-2016 by oral feeding successfully reproduced clear clinical symptoms, including vomiting, dehydration, and severe diarrhea in piglets. Collectively, these findings suggested that PDCoV was present in Guangdong, China and is highly virulent in piglets. Therefore, the development of potential vaccines against emerging PDCoV strains is urgently required.
References
Chen F, Zhu Y, Wu M, Ku X, Yao L, He Q (2015a) Full-length genome characterization of Chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc 3:e01284
Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J (2015b) Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 482:51–59
Chen Q, Gauger PC, Stafne MR, Thomas JT, Madson DM, Huang H, Zheng Y, Li G, Zhang J (2016) Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J Gen Virol 97:1107–1121
Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S (2016) Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106
Doyle LP, Hutchings LM (1946) A transmissible gastroenteritis in pigs. J Am Vet Med Assoc 108:257–259
Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ (2015a) Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol 53:1537–1548
Hu X Jr, Li N Jr, Tian Z Jr, Yin X Jr, Qu L, Qu J (2015b) Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. BMC Vet Res 11:72
Hu H, Jung K, Vlasova AN, Saif LJ (2016) Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch Virol 161:3421–3434
Hull RN, Cherry WR, Weaver GW (1976) The origin and characteristics of a pig kidney cell strain, LLC-PK. In Vitro 12:670–677
Jeong YJ, Park SI, Hosmillo M, Shin DJ, Chun YH, Kim HJ, Kwon HJ, Kang SY, Woo SK, Park SJ, Kim GY, Kang MI, Cho KO (2009) Detection and molecular characterization of porcine group C rotaviruses in South Korea. Vet Microbiol 138:217–224
Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ (2015) Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic Pigs. Emerg Infect Dis 21:650–654
Kong Q, Xue C, Ren X, Zhang C, Li L, Shu D, Bi Y, Cao Y (2013) Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteomic Sci 8:29
Lee S, Lee C (2014) Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc 2:e01191
Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J (2015) Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. Mbio 6:e00064
Madson DM, Magstadt DR, Arruda PH, Hoang H, Sun D, Bower LP, Bhandari M, Burrough ER, Gauger PC, Pillatzki AE, Stevenson GW, Wilberts BL, Brodie J, Harmon KM, Wang C, Main RG, Zhang J, Yoon KJ (2014) Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet Microbiol 174:60–68
Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, Pesko KN, Boutwell CL, Devincenzo JP, Ebel GD, Allen TM, Zody MC, Henn MR, Levin JZ (2013) Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res 41:e13
Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A (2014) Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infec Dis 20:1347–1350
Pensaert MB, De Bouck PD (1978) A new coronavirus-like particle associated with diarrhea in swine. Arch Virol 58:243–247
Reed LJ, Muench H (1938) A simple method of estimating of fifty per cent endpoints. Am J Epidemiol 27:493–497
Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D (2017) Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis 64:3–10
Sasseville AM, Boutin M, Gélinas AM, Dea S (2002) Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J Gen Virol 83:2411–2416
Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y (2015) Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis 62:575–580
Su M, Li C, Guo D, Wei S, Wang X, Geng Y, Yao S, Gao J, Wang E, Zhao X, Wang Z, Wang J, Wu R, Feng L, Sun D (2016) A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbentassay to detect antibodies against porcine deltacoronavirus. J Vet Med Sci 78:601–606
Wang L, Byrum B, Zhang Y (2014a) Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227–1230
Wang L, Byrum B, Zhang Y (2014b) Porcine coronavirus HKU15 detected in 9 US States, 2014. Emerg Infect Dis 20:1594–1595
Wang YW, Yue H, Fang W, Huang YW (2015) Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc 3:e00945
Wesley RD, Woods RD, Hill HT, Biwer JD (1990) Evidence for a porcine respiratory coronavirus, antigenically similar to transmissible gastroenteritis virus, in the United States. J Vet Diagn Invest 2:312–317
Woo PCY, Yi H, Lau SKP, Kwok-Yung Y (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2:1804–1820
Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY (2012) Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008
Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, Zhang H, Lv DH, Li F, Wang D (2016) Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J 13:136
Zhang J (2016) Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res 226:71–84
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0500101).
Author information
Authors and Affiliations
Contributions
YCC and ZCX conceived and designed the experiments; ZCX and HLZ performed the experiments; ZCX and HLZ analyzed the data; YCC, QFZ, YPD, LC, YZ and CYX contributed reagents/materials/analysis tools; ZCX wrote the paper. YCC checked and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The animal study was supervised by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (IACUC DD-17-0403) and used in accordance with regulation and guidelines of this committee.
Rights and permissions
About this article
Cite this article
Xu, Z., Zhong, H., Zhou, Q. et al. A Highly Pathogenic Strain of Porcine Deltacoronavirus Caused Watery Diarrhea in Newborn Piglets. Virol. Sin. 33, 131–141 (2018). https://doi.org/10.1007/s12250-018-0003-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-018-0003-8