Abstract
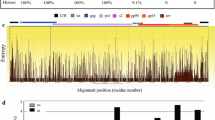
The envelope protein (Env) of lentiviruses such as HIV, SIV, FIV and EIAV is larger than that of other retroviruses. The Chinese EIAV attenuated vaccine is based on Env and has helped to successfully control this virus, demonstrating that envelope is crucial for vaccine. We compared Env variation of the four kinds of lentiviruses. Phylogenetic analysis showed that the evolutionary relationship of Env between HIV and SIV was the closest and they appeared to descend from a common ancestor, and the relationship of HIV and EIAV was the furthest. EIAV had the shortest Env length and the least number of potential N-linked glycosylation sites (PNGS) as well as glycosylation density compared to various immunodeficiency viruses. However, HIV had the longest Env length and the most PNGS. Moreover, the alignment of HIV and SIV showed that PNGS were primarily distributed within extracellular membrane protein gp120 rather than transmembrane gp41. It implies that the size difference among these viruses is associated with a lentivirus specific function and also the diversity of env. There are low levels of modification of glycosylation sites of Env and selection of optimal protective epitopes might be useful for development of an effective vaccine against HIV/AIDS.
Similar content being viewed by others
References
Bogers W M, Cheng-Mayer C, Montelaro R C. 2000. Developments in preclinical aids vaccine efficacy models. AIDS, 14 Suppl 3: S141–S151.
Burton D R, Desrosiers R C, Doms R W, et al. 2004. Hiv vaccine design and the neutralizing antibody problem. Nat Immunol, 5(3): 233–236.
Cavarelli M, Karlsson I, Zanchetta M, et al. 2008. Hiv-1 with multiple ccr5/cxcr4 chimeric receptor use is predictive of immunological failure in infected children. PLoS One, 3(9): e3292.
Clements J E, Zink M C. 1996. Molecular biology and pathogenesis of animal lentivirus infections. Clin Microbiol Rev, 9(1): 100–117.
Craigo J K, Zhang B, Barnes S, et al. 2007. Envelope variation as a primary determinant of lentiviral vaccine efficacy. Proc Natl Acad Sci U S A, 104(38): 15105–15110.
Desrosiers R C. 2004. Prospects for an aids vaccine. Nat Med, 10(3): 221–223.
Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol, 266: 418–427.
Johnson W E, Sanford H, Schwall L, et al. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J Virol, 77(18): 9993–10003.
Kuiken C, Korber B, Shafer R W. 2003. Hiv sequence databases. AIDS Rev, 5(1): 52–61.
Leroux C, Cadore J L, Montelaro R C. 2004. Equine infectious anemia virus (eiav): What has hiv’s country cousin got to tell us? Vet Res, 35(4): 485–512.
Liang H, He X, Shen R X, et al. 2006. Combined amino acid mutations occurring in the envelope closely correlate with pathogenicity of eiav. Arch Virol, 151(7): 1387–1403.
Losman B, Bolmstedt A, Schonning K, et al. 2001. Protection of neutralization epitopes in the v3 loop of oligomeric human immunodeficiency virus type 1 glycoprotein 120 by n-linked oligosaccharides in the v1 region. AIDS Res Hum Retroviruses, 17(11): 1067–1076.
Losman B, Biller M, Olofsson S, et al. 1999. The n-linked glycan of the v3 region of hiv-1 gp120 and cxcr4-dependent multiplication of a human immunodeficiency virus type 1 lymphocyte-tropic variant. FEBS Lett, 454(1–2): 47–52.
Means R E, Greenough T, Desrosiers R C. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol, 71(10): 7895–7902.
Mellquist J L, Bowman B, Kasturi L, et al. 1998. Characterization of hiv type 1 gp120 v3 region sequences from ugandan infants. AIDS Res Hum Retroviruses, 14(15): 1391–1395.
Miyauchi K, Curran A R, Long Y, et al. The membrane-spanning domain of gp41 plays a critical role in intracellular trafficking of the hiv envelope protein. Retrovirology, 7: 95.
Mori K, Yasutomi Y, Ohgimoto S, et al. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus sivmac239 in rhesus macaques: Robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J Virol, 75(9): 4023–4028.
Mori K, Sugimoto C, Ohgimoto S, et al. 2005. Influence of glycosylation on the efficacy of an env-based vaccine against simian immunodeficiency virus sivmac239 in a macaque aids model. J Virol, 79(16): 10386–10396.
Ohgimoto S, Shioda T, Mori K, et al. 1998. Location-specific, unequal contribution of the n glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol, 72(10): 8365–8370.
Payne S L, Pei X F, Jia B, et al. 2004. Influence of long terminal repeat and env on the virulence phenotype of equine infectious anemia virus. J Virol, 78(5): 2478–2485.
Reitter J N, Means R E, Desrosiers R C. 1998. A role for carbohydrates in immune evasion in aids. Nat Med, 4(6): 679–684.
Rice N R, Henderson L E, Sowder R C, et al. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol, 64(8): 3770–3778.
Schonning K, Jansson B, Olofsson S, et al. 1996. Rapid selection for an n-linked oligosaccharide by monoclonal antibodies directed against the v3 loop of human immunodeficiency virus type 1. J Gen Virol, 77 ( Pt 4): 753–758.
Sellon D C, Fuller F J, McGuire T C. 1994. The immunopathogenesis of equine infectious anemia virus. Virus Res, 32(2): 111–138.
Shen R X, Xu Z D, HE Y S, et al. 1979. Development of a eiav donkey leucocyte attenuated vaccine. Sci Agr Sin, 4: 1–15. (in Chinese)
Shen T, Liang H, Tong X, et al. 2006. Amino acid mutations of the infectious clone from chinese eiav attenuated vaccine resulted in reversion of virulence. Vaccine, 24(6): 738–749.
Sjolander S, Bolmstedt A, Akerblom L, et al. 1996. N-linked glycans in the cd4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of t cells recognizing an epitope located in their vicinity. Virology, 215(2): 124–133.
Sober E. 2004. The contest between parsimony and likelihood. Syst Biol, 53(4): 644–653.
Thompson J D, Gibson T J, Plewniak F, et al. 1997. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res, 25(24): 4876–4882.
Tully D C, Wood C. 2010. Chronology and evolution of the hiv-1 subtype c epidemic in ethiopia. AIDS, 24(10): 1577–1582.
Zhang H, Tully D C, Hoffmann F G, et al. 2010. Restricted genetic diversity of hiv-1 subtype c envelope glycoprotein from perinatally infected zambian infants. PLoS One, 5(2): e9294.
Zhang M, Gaschen B, Blay W, et al. 2004. Tracking global patterns of n-linked glycosylation site variation in highly variable viral glycoproteins: Hiv, siv, and hcv envelopes and influenza hemagglutinin. Glycobiology, 14(12): 1229–1246.
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation items: Natural Science Foundation of China (30970162) and Tianjin Municipal Science and Technology Foundation (08ZCGHHZ01800).
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yuan, Q., Liu, C., Liang, Z. et al. The comparison of genetic variation in the envelope protein between various immunodeficiency viruses and equine infectious anemia virus. Virol. Sin. 27, 241–247 (2012). https://doi.org/10.1007/s12250-012-3253-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-012-3253-x