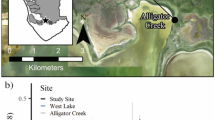
Abstract
In response to current threats to salt marshes, there are increasing efforts to restore these vital coastal ecosystems and promote their resilience to global change drivers. Unfortunately, the economic cost associated with assessing the effectiveness of restoration is prohibitive and more information is needed about the trajectory and timing of restoration outcomes to improve restoration practices. Microbial communities provide essential salt marsh functions so assessing the degree to which microbial communities in restored marshes resemble reference marshes can serve as a proxy indicator for the potential return of microbial function. We studied a recently restored marsh located on Cape Cod, MA, USA, by examining shifts in the microbial community and sediment edaphic properties in three habitats of a degraded oligohaline marsh, both before and after restoration of tidal flooding and in comparison with three nearby S. alterniflora reference marshes that never had flow restrictions. We hypothesized that the microbial community would respond rapidly to the restoration and that over time these communities would be indistinguishable from reference marsh communities. We found that soil edaphic characteristics shifted along a trajectory of recovery toward the reference marshes, with increases in salinity and decreases in soil organic matter, percentage of carbon, and percentage of nitrogen. The microbial communities in all three habitats within the restored marsh were different from reference marshes, and both the prokaryotic and fungal communities within P. australis and Typha sp. habitats became more similar to reference marshes (similarities increasing from an average of 5 to 16% for prokaryotes and 3 to 10% for fungi) during the first 2 years after restoration. In that same time period, by contrast, there was no return of the native marsh vegetation. These results suggest that shifts in microbial community structure occur prior to shifts in marsh vegetation and may facilitate the successful revegetation of restored marshes. Understanding the recovery trajectory of marshes during restoration and the role that microbes play in promoting the long-term sustainability of these habitats is essential; these results suggest that microbial communities respond rapidly and in a positive direction to restoration efforts.







Similar content being viewed by others
References
Able, K.W., and S.M. Hagan. 2003. Impact of common reed, Phragmites australis, on essential fish habitat: Influence on reproduction, embryological development, and larval abundance of Mummichog (Fundulus heteroclitus). Estuaries 26: 40–50.
Anderson, M. J. 2017. Permutational multivariate analysis of variance (PERMANOVA), Wiley StatsRef: Statistics Reference Online. https://doi.org/10.1002/9781118445112.stat07841.
Barberán, A., L. Mcguire, J.A. Wolf, F.A. Jones, S.J. Wright, B.L. Turner, A. Essene, S.P. Hubbell, B.C. Faircloth, and N. Fierer. 2015. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecology Letters 18 (12): 1397–1405.
Bates, D., M. Mächler, B. Bolker, and S. Walker. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. https://doi.org/10.18637/jss.v067.i01.
Bernhard, A.E., D. Marshal, and L. Yiannos. 2012. Increased variability of microbial communities in restored marshes nearly 30 years after tidal flow restoration. Estuaries and Coasts 35: 1049–1059.
Bernhard, A.E., C. Dwyer, A. Idrizi, G. Bender, and R. Zwick. 2015. Long-term impacts of disturbance on nitrogen cycling bacteria in a New England salt marsh. Fronteirs in Microbiology. https://doi.org/10.3389/fmicb.2015.00046.
Bowen, J.L., H.G. Morrison, J.E. Hobbie, and M.L. Sogin. 2012. Salt marsh sediment diversity: A test of the variability of the rare biosphere among environmental replicates. The ISME Journal 6 (11): 2014–2023.
Bowen, J.L., D. Weisman, M. Yasuda, A. Jayakumar, H.G. Morrison, and B.B. Ward. 2015. Marine oxygen-deficient zones harbor depauperate denitrifying communities compared to novel genetic diversity in coastal sediments. Microbial Ecology 70: 311–321.
Bowen, J.L., P.J. Kearns, J.E.K. Byrnes, S. Wigginton, W.J. Allen, M. Greenwood, K. Tran, J. Yu, J.T. Cronin, and L.A. Meyerson. 2017. Lineage overwhelms environmental conditions in determining rhizosphere bacterial community strucutre in a cosmopolitan invasvive plant. Nature Communications 8: 433.
Broeckling, C.D., A.K. Broz, J. Bergelson, D.K. Manter, and J.M. Vivanco. 2008. Root exudates regulate soil fungal community composition and diversity. Applied and Environmental Microbiology 74 (3): 738–744.
Buchan, A., S.Y. Newell, M. Butler, E.J. Biers, J.T. Hollibaugh, and M.A. Moran. 2003. Dynamics of bacterial and fungal communities on decaying salt marsh grass. Applied and Environmental Microbiology 69 (11): 6676–6687.
Burke, D., E. Hamerylynck, and D. Hahn. 2002. Interactions among plant species and microorganisms in salt marsh sediments. Applied and Environmental Microbiology 68 (3): 1157–1164.
Callahan, B.J., P.J. McMurdie, M.J. Rosen, A.W. Han, A.J.A. Johnson, and S.P. Holmes. 2016. DADA2: High resolution sample inference from Illumina amplicon data. Nature Methods 13 (7): 581–583.
Callahan, B.J., P.J. McMurdie, and S.P. Holmes. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME Journal 11 (12): 2639–2643.
Cao, Y., P.G. Green, and P.A. Holden. 2008. Microbial community composition and denitrifying enzyme activities in salt marsh sediments. Applied and Environmental Microbiology 74 (24): 7585–7595.
Cape Cod Conservation District. 2012. Final technical memorandum Muddy Creek Wetland Restoration Chatham, and Harwich, Massachusetts. Providence: Fuss and O'Neill.
Caporaso, J.G., J. Kuczynski, J. Strombaugh, K. Bittinger, F.D. Bushman, E.K. Costello, et al. 2010. QIIME allows analysis of high-throughput community seuqencing data. Nature Methods 7: 335–336.
Caporaso, J.G., C.L. Lauber, W.A. Walters, D. Berg-Lyons, C.A. Lozupone, P.J. Turnbaugh, et al. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences 108: 4516–4522.
Caporaso, J.G., C.L. Lauber, W.A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S.M. Owens, J. Betley, L. Fraser, M. Bauer, N. Gormley, J.A. Gilbert, G. Smith, and R. Knight. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal 6 (8): 1621–1624.
Chen, Y.L., T.L. Xu, S.D. Veresoglou, H.W. Hu, Z.P. Hao, Y.J. Hu, L. Liu, Y. Deng, M.C. Rillig, and B.D. Chen. 2017. Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biology and Biochemistry 110: 12–21.
Costanza, R., S.C. Farber, and J. Maxwell. 1989. Valuation and management of wetland ecosystems. Ecological Economics 1: 335–361.
Craft, C., P. Megonigal, S. Broome, J. Stevenson, R. Freese, J. Cornell, L. Zheng, and J. Sacco. 2003. The pace of ecosystem development of constructed Spartina alterniflora marshes. Ecological Applications 13 (5): 1417–1432.
Crump, B.C., C.S. Hopkinson, M.L. Sogin, and J.E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: Combined influences of bacterial growth and residence time. Applied and Environmental Microbiology 70 (3): 1494–1505.
Csardi, G., and T. Nepusz. 2006. The igraph software package for complex network reseach. InterJournal Complex Systems 1695: 1–9.
Daleo, P., J. Alberti, A. Canepuccia, M. Escapa, E. Fanjul, B.R. Silliman, M.D. Bertness, and O. Iribarne. 2008. Mycorrhizal fungi determine salt-marsh plant zonation depending on nutrient supply. Journal of Ecology 96: 431–437.
de León, D.G., M. Moora, M. Öpik, L. Neuenkamp, M. Gerz, T. Jairus, M. Vasar, C.G. Bueno, J. Davison, and M. Zobel. 2016. Symbiont dynamics during ecosystem succession: Co-occurring plant and arbuscular mycorrhizal fungal communities. FEMS Microbiology Ecology 92: 1–9.
Delgado-Baquerizo, M., F.T. Maestre, P.B. Reich, T.C. Jeffries, J.J. Gaitan, et al. 2016. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nature Communications. https://doi.org/10.1038/ncomms10541.
Dini-Andreote, F., M.J. de Brossi, J.D. van Elsas, and J.F. Salles. 2016a. Reconstructing the genetic potential of the microbially-mediated nitrogen cycle in a salt marsh ecosystem. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2016.00902.
Dini-Andreote, F., V.S. Pylro, P. Baldrian, J.D. Van Elsas, and J.F. Salles. 2016b. Ecological succession reveals potential signatures of marine-terrestrial transition in salt marsh fungal communities. The ISME Journal 10: 1984–1997.
Duarte, B., J. Freitas, and I. Caçador. 2012. Sediment microbial activities and phsic-chemistry as progress indicators of salt marsh restoration success. Ecological Indicators 19: 231–239.
Erlacher, A., T. Cernava, M. Cardinale, J. Soh, C.W. Sensen, M. Grube, and G. Berg. 2015. Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2015.00053.
Falkowski, P.G., T. Fenchel, and E.F. Delong. 2008. The microbial engines that drive Earth’s biogeochemical cycles. Science 320 (5879): 1034–1039.
Fierer, N. 2017. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nature Reviews Microbiology 15: 579–590.
Fierer, N., and J.T. Lennon. 2011. The generation and maintenance of diversity in microbial communities. American Journal of Botany 98 (3): 439–448.
Hamilton, E.W., and D.A. Frank. 2001. Can plants stimulate soil microbes and their own nutrient supply? Evidence from a grazing tolerant grass. Ecology 82: 2397–2402.
Hartman, W.H., C.J. Richardson, R. Vilgalys, and G.L. Bruland. 2008. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proceedings of the National Academy of Sciences 105: 17842–17847.
Hartmann, A., M. Schmid, D. van Tuinen, and G. Berg. 2009. Plant-driven selection of microbes. Plant and Soil 321: 235–257.
Hothorn, T., F. Bretz, and P. Westfall. 2008. Simultaeous inference in general parametric models. Biometrical Journal 50 (3): 346–363.
Howarth, R.W., and A.E. Giblin. 1983. Sulfate reduction in the salt marshes at Sapelo Island, Georgia. Limnology and Oceanography 28: 70–82.
Ivanova, E.P., S. Flavier, and R. Christen. 2004. Phylogenetic relationships among marine Alteromonas-like proteobacteria: Emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferri. International Journal of Systematic and Evolutionary Microbiology 54: 1773–1788.
Liang, B., L.Y. Wang, S.M. Mbadinga, J.F. Liu, S.Z. Uang, J.D. Gu, and B.Z. My. 2015. Anaerolineaceae and Methanosaeta turned to be the dominant microorgnaisms in alkanes-dependent mathanogenic culture after long-term incubation. AMB Express. https://doi.org/10.1186/s13568-015-0117-4.
Liaw, A., and M. Wiener. 2002. Classification and regression by randomForest. Rnews 2: 18–22.
Lindström, E.S., M.P.K. Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Applied and Environmental Microbiology 71 (12): 8201–8206.
Liu, Z., N.U. Frigaard, K. Vogl, T. Iino, M. Ohkuma, J. Overmann, and D.A. Byant. 2012. Complete genomie of Ignavibacterium album, a metabolically versatile, flaellated, facultative anaerobe from the phylum Chlorobi. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2012.00185.
Lozupone, C.A., and R. Knight. 2007. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences 104: 11436–11440.
Ma, Z., M. Zhang, R. Xiao, Y. Ciu, and F. Yu. 2017. Changes in soil microbial biomass and community composition in coastal wetlands affected by restoration projects in a Chinese delta. Geoderma 289: 124–134.
McDonald, D., M.N. Price, J. Goodrich, E.P. Nawrocki, T.Z. Desantis, A. Probst, et al. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal 6 (3): 610–618.
McHugh, J.M., and J. Dighton. 2004. Influence of mycorrhizal inoculation, inundation period, salinity, and phosphorus availability on the growth of two salt marsh grasses, Spartina alterniflora Lois. and Spartina cynosuroides (L.) Roth., in nursery systems. Restoration Ecology 12: 533–545.
McLeod, E., G.L. Chmura, S. Bouillon, R. Salm, M. Björk, C.M. Duarte, et al. 2011. A blueprint for blue carbon: Toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Frontiers in Ecology and the Environment 9: 552–560.
McMurdie, P.J., and S. Holmes. 2013. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. https://doi.org/10.1371/journal.pone.0061217.
Meyerson, L.A., A.M. Lambert, and K. Saltonstall. 2010. A tale of three lineages: Expansion of common reed (Phragmites australis) in the US Southwest and Gulf Coast. Invasive Plant Science and Management 3: 515–520.
Millard, P., and B.K. Singh. 2010. Does grassland vegetation drive soil microbial diversity? Nutrient Cycling in Agroecosystems 88: 147–158.
Moore, G.E., D.M. Burdick, C.R. Peter, and R. Donald. 2016. Belowground biomass of Phragmites australis in coastal marshes. Northeastern Naturalist 19: 611–626.
National Academies of Sciences, Engineering, and Medicine. 2017. Effective monitoring to evaluate ecological restoration in the Gulf of Mexico. Washington, DC: The National Academies Press. https://doi.org/10.17226/23476.
Neubauer, S.C. 2013. Ecosystem responses of a tidal freshwater marsh expereinceing saltwater intrusion and altered hydrology. Estuaries and Coasts 36: 491–507.
Newell, S.Y. 2001. Multiyear patterns of fungal biomass dynamics and productivity within naturally decaying smooth cordgrass shoots. Limnology and Oceanography 46: 573–583.
Newman, M.E. 2006. Finding community structure in networks using the eigenvectors of matricies. Physical Review E. https://doi.org/10.1103/PhysRevE.74.036104.
Nilsson, R.H., K.-H. Larsson, A.F.S. Taylor, J. Bengtsson-Palme, T.S. Feppesen, D. Schigel, et al. 2018. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 1: gky1022.
Oksanen, J., C. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, et al. 2018. vegan: Community Ecology Package R package version 2.5-2.
Oren, A. 2014. The family Xanthobacteraceae. In The prokaryotes, ed. E. Rosenberg, E.F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, 709–726. Berlin: Springer.
Pennings, S.C., and M.D. Bertness. 2001. Salt marsh communities. In Marine community ecology, ed. M.D. Bertness, S.D. Haines, and M. Hay, 289–316. Sunderland: Sinaur Associates.
Piehler, M.F., C.A. Currin, R. Cassanova, and H.W. Paerl. 1998. Development and N2-fixing activity of the benthic microbial community in transplanted Spartina alterniflora marshes in North Carolina. Restoration Ecology 6: 290–296.
Pjevac, P., M. Korlević, J.S. Berg, E. Bura-Nakić, I. Ciglenečki, R. Amann, and S. Orlić. 2015. Community shift from phototrophic to chemotrophic sulfide oxidation following anoxic holomixis in a stratified seawater lake. Applied and Environmental Microbiology 81 (1): 298–308.
Portnoy, J.W., and A.E. Giblin. 1997. Effects of historic tidal restrictions on salt marsh sediment chemistry. Biogeochemistry 36: 275–303.
Prober, S.M., J.W. Leff, S.T. Bates, E.T. Borer, J. Firn, W.S. Harpole, et al. 2015. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecology Letters 18: 85–95.
R Core Team. 2017. R: A Language and Environment for Statistical Computing. https://www.R-project.org/
Ramirez, K.S., J.W. Leff, A. Barberán, S.T. Bates, J. Betley, T.W. Crowther, et al. 2014. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proceedings of the Royal Society B. https://doi.org/10.1098/rspb.2014.1988.
Rickey, M.A., and R.C. Anderson. 2004. Effects of nitrogen addition on the invasive grass Phragmites australis and a native competitor Spartina pectinata. Journal of Applied Ecology 41: 888–896.
Rognes, T., T. Flouri, B. Nichols, C. Quince, and F. Mahé. 2016. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4: e2584.
Roman, C.T., and D.M. Burdick. 2012. A synthesis of research and practice on restoring tides to salt marshes. In Tidal marsh restoration: A synthesis of science and management, ed. C.T. Roman and D.M. Burdick, 2–10. Washington DC: Island Press.
Rublee, P., and B.E. Dornseif. 1978. Direct counts of bacteria in the sediments of a North Carolina salt marsh. Estuaries 1: 3–6.
Ruiz-Jaen, M.C., and T.M. Aide. 2005. Restoration success: How is it being measured? Restoration Ecology 13: 569–577.
Santín, C., J.M. de la Rosa, H. Knicker, X.L. Otero, M.Á. Álvarez, and F.J. González-Vila. 2009. Effects of reclamation and regeneration processes on organic matter from estuarine soils and sediments. Organic Geochemistry 40: 931–941.
Shange, R.S., R.O. Ankumah, A.M. Ibekwe, R. Zabawa, and S.E. Dowd. 2012. Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PLoS One. https://doi.org/10.1371/journal.pone.0040338.
Sinicrope, T.L., P.G. Hine, R.S. Warren, and W.A. Niering. 1990. Restoration of an impounded salt marsh in New England. Estuaries 13: 25–30.
Smith, S.M., and R.S. Warren. 2012. Vegetation responses to tidal restoration. In Tidal marsh restoration: A synthesis of science and management, ed. C.T. Roman and D.M. Burdick, 59–80. Washington DC: Island Press.
Smith, S.M., C.T. Roman, M.J. James-Pirri, K. Chapman, J. Portnoy, and E. Gwilliam. 2009. Responses of plant communities to incremental hydrologic restoration of a tide-restricted salt marsh in southern New England (Massacusetts, U.S.A.). Restoration Ecology 17: 606–618.
Tedersoo, L., M. Bahram, S. Põlme, U. Kõljalg, N.S. Yorou, K. Abarenkov, et al. 2014. Global diversity and geography of soil fungi. Science 346: 1052–1053.
Underwood, A.J. 1992. Beyond BACI: The detection of environmental impacts on populations in the real but variable world. Journal of Experimental Marine Biology and Ecology 161: 145–178.
USGS. 2018. National Water Information System. https://nwis.waterdata.usgs.gov/ma/nwis, Accessed: March, 2018.
Valiela, I., and M.L. Cole. 2002. Comparative evidence that salt marshes and mangroves may protect seagrass meadows from land-derived nitrogen loads. Ecosystems 5: 92–100.
Van Gemerden, H., and J. Mas. 1995. Ecology of phototrophic sulfur bacteria. In Anoxygenic photosynthetic bacteria. Advances in photosynthesis and respiration, ed. R.E. Blankenship, M.T. Madigan, and C.E. Bauer, 49–85. Dordrecht: Springer.
Waldrop, M.P., and M.K. Firestone. 2006. Response of microbial community composition and function to soil climate change. Microbial Ecology 52 (4): 716–724.
Walters, W., E.R. Hyde, D. Berg-lyons, G. Ackermann, G. Humphrey, A. Parada, J.A. Gilbert, and J.K. Jansson. 2015. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1: e0009–e0015.
Warren, R.S., P.E. Fell, R. Rozsa, A.H. Brawley, A.C. Orsted, E.T. Olson, V. Swamy, and W.A. Niering. 2002. Salt marsh restoration in Connecticut: 20 years of science and management. Restoration Ecology 10: 49–513.
Wickham, H. 2016. ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag.
Wortley, L., J.M. Hero, and M. Howes. 2013. Evaluating ecological restoration success: A review of the literature. Restoration Ecology 21: 537–543.
Yamamoto, K., Y. Shiwa, T. Ishige, H. Sakamoto, K. Tanaka, M. Uchino, N. Tanaka, S. Oguri, H. Saitoh, and S. Tsushima. 2018. Bacterial diveristy associate with the rhizosphere and endoshpere of two halophytes Glaux maritima and Salicornia europaea. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2018.02878.
Yao, Z., S. Du, C. Liang, Y. Zhao, F. Dini-Andreote, K. Wang, and D. Zhang. 2019. Bacterial community assembly in a typical estuarine marsh with mutliple environmental gradients. Applied and Environmental Microbiology. https://doi.org/10.1128/AEM.02602-18.
Zak, D.R., W.E. Holmes, D.C. White, A.D. Peacock, and D. Tilman. 2003. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 84: 2042–2050.
Zeng, Y., J. Baumbach, E.G. Barbosa, V. Azevedo, C. Zhang, and M. Koblížek. 2016. Metagenomic evidence for the presence of phototrophic Gemmatimonadetes bacteria in diverse environments. Envonmental Microbiology Reports 8: 139–149.
Acknowledgments
We would like to thank Georgeanne Keer and others at the Massachusetts Division of Ecological Restoration for sharing restoration plans and homeowners who allowed access to the marshes used in this study. Field assistance was provided by Annie Murphy and Itxaso Garay. Thanks are due to Alan Stebbins at University of Massachusetts Boston Environmental Analytical Facility (NSF 09-42371 and DBI:MRI-RI2 to Robyn Hannigan and Alan Christian) for assistance in the laboratory. Tom Goodkind, in the University of Massachusetts Boston Machine Shop, provided essential help in constructing sampling equipment associated with this experiment, and resources purchased with funds from the NSF FMSL program (DBI 1722553, to Northeastern University) were used to generate data for this manuscript.
Funding
This work was funded by an NSF CAREER grant (DEB1350491) to JLB.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Deana Erdner
Electronic supplementary material
ESM 1
(DOCX 942 kb)
Rights and permissions
About this article
Cite this article
Lynum, C.A., Bulseco, A.N., Dunphy, C.M. et al. Microbial Community Response to a Passive Salt Marsh Restoration. Estuaries and Coasts 43, 1439–1455 (2020). https://doi.org/10.1007/s12237-020-00719-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-020-00719-y