Abstract
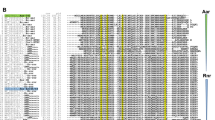
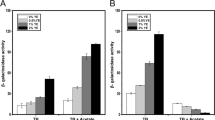
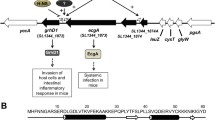
Enteropathogenic Escherichia coli (EPEC) is a diarrheagenic bacterium that predominantly infects infants in developing countries. EPEC forms attaching and effacing (A/E) lesions on the apical surface of the small intestine, leading to diarrhea. The locus of enterocyte effacement (LEE) is both necessary and sufficient for A/E lesion morphogenesis by EPEC. Gene expression from this virulence determinant is controlled by an elaborate regulatory web that extends beyond protein-based transcriptional regulators and includes small regulatory RNA (sRNA) that exert their effects posttranscriptionally. To date, only 4 Hfq-dependent sRNAs—MgrR, RyhB, McaS, and Spot42—have been identified that affect the LEE of EPEC by diverse mechanisms and elicit varying regulatory outcomes. In this study, we demonstrate that the paralogous Hfq-dependent sRNAs OmrA and OmrB globally silence the LEE to diminish the ability of EPEC to form A/E lesions. Interestingly, OmrA and OmrB do not appear to directly target a LEE-encoded gene; rather, they repress transcription from the LEE1 promoter indirectly, by means of an as-yet-unidentified transcriptional factor that binds within 200 base pairs upstream of the transcription start site to reduce the expression of the LEE master regulator Ler, which, in turn, leads to reduced morphogenesis of A/E lesions. Additionally, OmrA and OmrB also repress motility in EPEC by targeting the 5′ UTR of the flagellar master regulator, flhD.





Similar content being viewed by others
Data availability
The authors confirm that all relevant data are included in the article and/or its supplementary information files. All plasmids, strains, reagents, and other accessory materials will be expeditiously provided by the corresponding author, upon request, provided that the requestors pay for the shipping, packaging, and processing of samples.
References
Bak G, Lee J, Suk S et al (2015) Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci Rep 5:15287
Barba J, Bustamante VH, Flores-Valdez MA et al (2005) A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol 187:7918–7930
Bhatt S, Edwards AN, Nguyen HT et al (2009) The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun 77:3552–3568
Bhatt S, Romeo T, Kalman D (2011) Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol 19:217–224
Bhatt S, Egan M, Jenkins V et al (2016) The tip of the iceberg: on the roles of regulatory small RNAs in the virulence of enterohemorrhagic and enteropathogenic Escherichia coli. Front Cell Infect Microbiol 6:1–6
Bhatt S, Egan M, Ramirez J et al (2017a) Hfq and three Hfq-dependent small regulatory RNAs-MgrR, RyhB and McaS-coregulate the locus of enterocyte effacement in enteropathogenic Escherichia coli. Pathog Dis 75:1–14
Bhatt S, Jenkins V, Mason E et al (2017b) The small regulatory RNA Spot42 inhibits indole biosynthesis to negatively regulate the locus of enterocyte effacement of enteropathogenic Escherichia coli. Microorganisms 5:1–12
Brosse A, Korobeinikova A, Gottesman S et al (2016) Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res 44:9650–9666
Bustamante VH, Santana FJ, Calva E et al (2001) Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol 39:664–678
Bustamante VH, Villalba MI, Garcia-Angulo VA et al (2011) PerC and GrlA independently regulate Ler expression in enteropathogenic Escherichia coli. Mol Microbiol 82:398–415
Croxen MA, Finlay BB (2010) Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38
Datta S, Costantino N, Court DL (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115
De Lay N, Gottesman S (2012) A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538
De Lay N, Schu DJ, Gottesman S (2013) Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288:7996–8003
Deng W, Puente JL, Gruenheid S et al (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA 101:3597–3602
Elliott SJ, Wainwright LA, McDaniel TK et al (1998) The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 28:1–4
Friedberg D, Umanski T, Fang Y et al (1999) Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol 34:941–952
Gomez-Duarte OG, Kaper JB (1995) A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun 63:1767–1776
Guillier M, Gottesman S (2006) Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59:231–247
Guillier M, Gottesman S, Storz G (2006) Modulating the outer membrane with small RNAs. Genes Dev 20:2338–2348
Guillier M, Gottesman S (2008) The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36:6781–6794
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hansen AM, Kaper JB (2009) Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol Microbiol 73:446–465
Holmqvist E, Reimegard J, Sterk M et al (2010) Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J 29:1840–1850
Hu J, Torres AG (2015) Enteropathogenic Escherichia coli: foe or innocent bystander? Clin Microbiol Infect 21:729–734
Huang LH, Syu WJ (2008) GrlA of enterohemorrhagic Escherichia coli O157:H7 activates LEE1 by binding to the promoter region. J Microbiol Immunol Infect 41:9–16
Iguchi A, Thomson NR, Ogura Y et al (2009) Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol 191:347–354
Jarvis KG, Giron JA, Jerse AE et al (1995) Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA 92:7996–8000
Jobichen C, Li M, Yerushalmi G et al (2007) Structure of GrlR and the implication of its EDED motif in mediating the regulation of type III secretion system in EHEC. PLoS Pathog 3:e69
Lanata CF, Fischer-Walker CL, Olascoaga AC et al (2013) Global causes of diarrheal disease mortality in children < 5 years of age: a systematic review. PLoS ONE 8:e72788
Lenz DH, Mok KC, Lilley BN et al (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82
Link TM, Valentin-Hansen P, Brennan RG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA 106:19292–19297
Liu L, Johnson HL, Cousens S et al (2012) Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379:2151–2161
Liu X, Matsumura P (1994) The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176:7345–7351
Mandin P, Gottesman S (2009) A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72:551–565
Melamed S, Faigenbaum-Romm R, Peer A et al (2018) Mapping the small RNA interactome in bacteria using RIL-seq. Nat Protoc 13:1–33
Mellies JL, Elliott SJ, Sperandio V et al (1999) The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol 33:296–306
Mellies JL, Barron AM, Carmona AM (2007) Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75:4199–4210
Moller T, Franch T, Hojrup P et al (2002) Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9:23–30
Murphy KC, Campellone KG (2003) Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol 4:11
Padavannil A, Jobichen C, Mills E et al (2013) Structure of GrlR-GrlA complex that prevents GrlA activation of virulence genes. Nat Commun 4:2546
Papenfort K, Vogel J (2010) Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127
Platenkamp A, Mellies JL (2018) Environment controls LEE regulation in enteropathogenic Escherichia coli. Front Microbiol 9:1694
Centers for Disease Control and Prevention-United Stated (CDC-US) (2015) Diarrhea: common illness, global killer. https://stacks.cdc.gov/view/cdc/13557#tabs-2. Accessed 7 Dec 2012
Romilly C, Hoekzema M, Holmqvist E et al (2020) Small RNAs OmrA and OmrB promote class III flagellar gene expression by inhibiting the synthesis of anti-Sigma factor FlgM. RNA Biol 17:872–880
Sauder AB, Kendall MM (2021) A pathogen-specific sRNA influences enterohemorrhagic Escherichia coli fitness and virulence in part by direct interaction with the transcript encoding the ethanolamine utilization regulatory factor EutR. Nucleic Acids Res 49:10988–11004
Serapio-Palacios A, Finlay BB (2020) Dynamics of expression, secretion and translocation of type III effectors during enteropathogenic Escherichia coli infection. Curr Opin Microbiol 54:67–76
Shakhnovich EA, Davis BM, Waldor MK (2009) Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol 74:347–363
Sudo N, Soma A, Iyoda S et al (2018) Small RNA Esr41 inversely regulates expression of LEE and flagellar genes in enterohaemorrhagic Escherichia coli. Microbiology (reading) 164:821–834
Tauschek M, Yang J, Hocking D et al (2010) Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J Bacteriol 192:3722–3734
Umanski T, Rosenshine I, Friedberg D (2002) Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735–2744
Updegrove TB, Zhang A, Storz G (2016) Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138
Valentin‐Hansen P, Eriksen M, Udesen C (2004) The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol 51:1525–1533
Vogt SL, Raivio TL (2014) Hfq reduces envelope stress by controlling expression of envelope-localized proteins and protein complexes in enteropathogenic Escherichia coli. Mol Microbiol 92:681–697
Wang D, McAteer SP, Wawszczyk AB et al (2018) An RNA-dependent mechanism for transient expression of bacterial translocation filaments. Nucleic Acids Res 46:3366–3381
Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136:615–628
Zhang A, Wassarman KM, Ortega J et al (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell 9:11–22
Acknowledgements
S.B. is eternally grateful to Gigi Storz (NIH/NICHD), Dan Kalman (Emory University), and Chris Weingart (Denison University) for their continued support and mentoring throughout his career. Z.M. is a recipient of the SJU McNulty scholarship whereas S.M. and A.M. are recipients of the SJU McNulty Fellowship.
Funding
This research was supported by start-up funds generously provided by Saint Joseph’s University (SJU) with additional support provided by the SJU Biology department and the McNulty Foundation.
Author information
Authors and Affiliations
Contributions
SB conceived and designed the project. SB contributed the reagents. SM, JEF, AM, ZM, SC, MM, TB, TH, WY, BA, MR, and SB collected the data. SB performed the data analysis and wrote the paper.
Corresponding author
Ethics declarations
Research involving human participants and/or animals
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muche, S., El-Fenej, J., Mihaita, A. et al. The two sRNAs OmrA and OmrB indirectly repress transcription from the LEE1 promoter of enteropathogenic Escherichia coli. Folia Microbiol 68, 415–430 (2023). https://doi.org/10.1007/s12223-022-01025-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-01025-9