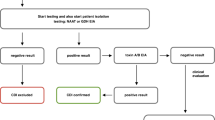
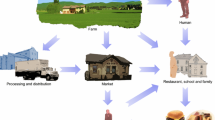
Abstract
Globally, it is estimated that there are 2 billion cases of diarrhoeal disease each year, with 525,000 children under the age of 5 years, dying from diarrhoea. This also affects 1 in 5 people in the UK each year. Rapid diagnosis, appropriate treatment and infection control measures are, therefore, particularly important. Currently, Public Health Wales and England Microbiology Division test for five key bacterial gastrointestinal pathogens, i.e. Escherichia coli O157 (VTEC), Shigella dysenteriae, Salmonella spp., Campylobacter spp. and Clostridioides difficile. There is, however, a poor success rate with identification of these pathogens, leaving the patient at risk from untreated infections. This study has developed effective and reliable tools with a high positive outcome for diagnosis of diarrhoeal infection. The study blindly analysed 592 samples, with the most abundant species being Shigella sonnei at 15%, and the top genus Bacteroides at 26%. Campylobacter spp. had an abundance of 4%, Clostridium difficile 3%, and Salmonella spp. 0.2%. There were also significant differences in abundance at genus level, between the Flemish Gut project and diarrhoea samples, with respect to Shigella (0.2%) and Campylobacter (0.1%). The project introduced a novel Shigella spp. (Escherichia) threshold of 5.32% to determine (Escherichia) a healthy or unhealthy community. A DMBiome model was developed to integrate the 5.32% threshold of Shigella spp., the Public Health laboratory tested pathogens, and two emerging enteropathogens. The overall positive outcome was that 89% of all samples were diagnosed with diarrhoea infections, leaving 11% unknown.





Similar content being viewed by others
Availability of data and material
Sequences are accessible under the study accession number of PRJEB35355 (https://www.ebi.ac.uk/ena).
References
Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP (2020) Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog 12:1. https://doi.org/10.1186/s13099-019-0341-6
Ammer-Herrmenau C, Pfisterer N, Weingarten MFJ, Neesse A (2020) The microbiome in pancreatic diseases: recent advances and future perspectives. United European Gastroenterol J 8:8. https://doi.org/10.1177/2050640620944720
Arasaradnam RP, Brown S, Forbes A et al (2018) Guidelines for the investigation of chronic diarrhoea in adults. British Society of Gastroenterology, 3rd edition. Gut 67:1380–1399. https://doi.org/10.1136/gutjnl-2017-315909
Bibbò S, Abbondio M, Sau R et al (2020) Fecal microbiota signatures in celiac disease patients with poly-autoimmunity. Front Cell Infect Microbiol 10:349. https://doi.org/10.3389/fcimb.2020.00349
Binder HJ (2009) Mechanisms of diarrhea in inflammatory bowel diseases. Ann N Y Acad Sci 1165:285–293. https://doi.org/10.1111/j.1749-6632.2009.04039.x
Brelian D, Tenner S (2012) Diarrhoea due to pancreatic diseases. Best Pract Res Clin Gastroenterol 26:623–631. https://doi.org/10.1016/j.bpg.2012.11.010
Clemente JC, Ursell LK, Parfrey LW, Knight R (2012) The impact of the gut microbiota on human health. An integrative view. Cell 148:1258–1270. https://doi.org/10.1016/j.cell.2012.01.035
Clooney AG, Eckenberger J, Laserna-Mendieta E et al (2020) Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut 2020:321106. https://doi.org/10.1136/gutjnl-2020-321106
Davis-Richardson AG, Ardissone AN, Dias R et al (2014) Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol 5:678. https://doi.org/10.3389/fmicb.2014.00678
Deng H, Yang S, Zhang Y et al (2019) Bacteroides fragilis prevents clostridium difficile infection in a mouse model by restoring gut barrier and microbiome regulation. Front Microbiol 10:601. https://doi.org/10.3389/fmicb.2019.00601
Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK (2008) Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J 2:716–727. https://doi.org/10.1038/ismej.2008.37
Dicpinigaitis PV, De Aguirre M, Divito J (2015) Enterococcus hirae Bacteremia associated with acute pancreatitis and septic shock. Case Reports in Infectious Diseases 123852:3. https://doi.org/10.1155/2015/123852
Ding RX, Goh WR, Wu RN, Yue XQ, Luo X, Khine WWT, Wu JR, Lee YK (2019) Revisit gut microbiota and its impact on human health and disease. J Food Drug Anal 27:623–631. https://doi.org/10.1016/j.jfda.2018.12.012
Dixon P (2013) VEGAN, a package of R functions for community ecology. Appl Veg Sci 14:927–930. https://doi.org/10.2307/3236992
Dong TS, Gupta A (2019) Influence of early life, diet, and the environment on the microbiome. Clin Gastroenterol Hepatol 17:231–242. https://doi.org/10.1016/j.cgh.2018.08.067
DuPont HL (2016) Persistent diarrhea: a clinical review. JAMA 315:2712–2723. https://doi.org/10.1001/jama.2016.7833
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638. https://doi.org/10.1126/science.1110591
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA (2011) NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745–757. https://doi.org/10.1016/j.cell.2011.04.022
Falony G, Joossens M, Vieira-Silva S et al (2016) Population-level analysis of gut microbiome variation. Science 352:560–564. https://doi.org/10.1126/science.aad3503
Hasan N, Yang H (2019) Factors affecting the composition of the gut microbiota, and its modulation. Peer J 7:e7502. https://doi.org/10.7717/peerj.7502
Hawrelak AJ, Myers SP (2004) The causes of intestinal dysbiosis: A review. Altern Med Rev 9:80–197 (PMID: 15253677)
Humphries RM, Linscott AJ (2015) Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. https://doi.org/10.1128/CMR.00073-14
Ignacio A, Fernandes MR, Avila-Campos MJ, Nakano V (2015) Enterotoxigenic and non-enterotoxigenic Bacteroides fragilis from fecal microbiota of children. Braz J Microbiol 46:1141–1145. https://doi.org/10.1590/S1517-838246420140728
Ilinskaya ON, Ulyanova VV, Yarullina DR, Gataullin IG (2017) Secretome of Intestinal Bacilli: a natural guard against pathologies. Front Microbiol 8:1666. https://doi.org/10.3389/fmicb.2017.01666
Joensen KG, Engsbro AL, Lukjancenko O, Kaas RS, Lund O, Westh H, Aarestrup FM (2017) Evaluating next-generation sequencing for direct clinical diagnostics in diarrhoeal disease. Eur J Clin Microbiol Infect Dis 36:1325–1338. https://doi.org/10.1007/s10096-017-2947-2
KaabiAL-Yassari HA (2019) 16SrRNA sequencing as tool for identification of Salmonella spp isolated from human diarrhea cases. J Phys: Conf Ser 1294:062041. https://doi.org/10.1088/1742-6596/1294/6/062041
Khanna S (2020) Management of Clostridioides difficile infection in patients with inflammatory bowel disease. Intest Res. https://doi.org/10.5217/ir.2020.00045
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Leo S, Lazarevic V, Gaïa N, Estellat C, Girard M, Matheron S, Armand-Lefèvre L, Andremont A (2019) The intestinal microbiota predisposes to traveler’s diarrhea and to the carriage of multidrug-resistant Enterobacteriaceae after traveling to tropical regions. Gut Microbes. https://doi.org/10.1080/19490976.2018.1564431
Liu H, Guo M, Jiang Y et al (2019) Diagnosing and tracing the pathogens of infantile infectious diarrhea by amplicon sequencing. Gut Pathog 11:12. https://doi.org/10.1186/s13099-019-0292-y
O’Brien CL, Kiely CJ, Pavli P (2018) The microbiome of Crohn’s disease aphthous ulcers. Gut Pathog 10:44. https://doi.org/10.1186/s13099-018-0265-6
Paczosa MK, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. https://doi.org/10.1128/MMBR.00078-15
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. https://doi.org/10.1093/bioinformatics/btu494
Perry MD, Corden SA, Howe RA (2014) Evaluation of the luminex xTAG gastrointestinal pathogen panel and the savyon diagnostics gastrointestinal infection panel for the detection of enteric pathogens in clinical samples. J Med Microbiol 63:1419–1426. https://doi.org/10.1099/jmm.0.074773-0
PHE (2014) UK standards for microbiology investigations: investigation of faecal specimens for enteric pathogens. Bacteriology B30: 8.1:1–41. https://www.gov.uk/government/publications/smi-b-30-investigation-of-faecal-specimens-for-enteric-pathogens. Assessed 07 November 2020
Platts-Mills JA, Liu J, Houpt ER (2013) New concepts in diagnostics for infectious diarrhea. Mucosal Immunol 6:876–885. https://doi.org/10.1038/mi.2013.50
Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C (2011) Gut microbiota: Next frontier in understanding human health and development of biotherapeutics. Biologics: Targets & Therapy V5:71–86. https://doi.org/10.2147/BTT.S19099
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Shane AL, Mody RK, Crump JA et al (2017) Infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 65:e45–e80. https://doi.org/10.1093/cid/cix669
Tam CC, O’Brien SJ (2016) Economic cost of Campylobacter, Norovirus and Rotavirus disease in the United Kingdom. PLoS One 11:e0138526. https://doi.org/10.1371/journal.pone.0138526
Tingting S, Rongbei L, Allen L et al (2018) Altered intestinal microbiota with increased abundance of Prevotella is associated with high risk of diarrhea-predominant irritable bowel syndrome. Gastroenterol Res Pract. https://doi.org/10.1155/2018/6961783
Vandeputte D, Tito R, Vanleeuwen R, Falony G, Raes J (2017) Practical considerations for large-scale gut microbiome studies. FEMS Microbiol Rev 41:154–167. https://doi.org/10.1093/femsre/fux027
Varela E, Manichanh C, Gallart M, Torrejon A, Borruel N, Casellas F, Guarner F, Antolin M (2013) Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther 38. https://doi.org/10.1111/apt.12365
Vidal R, Ginard D, Khorrami S et al (2015) Crohn associated microbial communities associated to colonic mucosal biopsies in patients of the western Mediterranea. Syst Appl Microbiol 38:442–452. https://doi.org/10.1016/j.syapm.2015.06.008
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
WHO (2017) Diarrhoeal disease. https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease. Accessed 09 October 2019
WHO (2020) Food safety- key facts. Publishing WHO. https://www.who.int/news-room/fact-sheets/detail/food-safety. Accessed 06 November 2020
Wick EC, Sears CL (2010) Bacteroides spp. and diarrhea. Curr Opin Infect Dis 23:470–474. https://doi.org/10.1097/QCO.0b013e32833da1eb
Wilson M, Martin R, Walk ST et al (2011) Clinical and laboratory features of Streptococcus salivarius meningitis. A case report and literature review. Clin Med Res 10:15–25. https://doi.org/10.3121/cmr.2011.1001
Ye Z, Huang Y, Zheng C et al (2019) Clinical and genetic spectrum of children with congenital diarrhea and enteropathy in China. Genet Med 21:2224–2230. https://doi.org/10.1038/s41436-019-0488-z
Zamani S, Hesam S, Zali MR et al (2017) Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog 9:53. https://doi.org/10.1186/s13099-017-0202-0
Zhang YJ, Li S, Gan GY, Zhou T, Xu DP, Li HB (2015) Impacts of gut bacteria on human health and diseases. Int J Mol Sci 16:7493–7519. https://doi.org/10.3390/ijms16047493
Zhou H, Zhao X, Sun L et al (2020) Gut microbiota profile in patients with type 1 diabetes based on 16S rRNA gene sequencing: a systematic review. Dis Markers 3936247. https://doi.org/10.1155/2020/3936247
Acknowledgements
I would like to thank Dr Julie Smith who proofread the paper.
Funding
The 592 samples used in this project was initially funded by BBSRC (under the FLIP programme [BB/J019143/1]) where the author was research co-applicant. The lack of data generated from the project did not address the project objectives and therefore could not be published. However, Public Health Wales and the Principle Investigator of the BBSRC grant gave the author permission to use the 592 samples.
Author information
Authors and Affiliations
Contributions
Dr Ann Smith analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Ethical approval
Ethical approval was not necessary as this project used raw samples from a previous study.
Consent for publication
I had consent from Public Health Wales to use the raw samples for my own research interest, and there were no patient and/or microbiological data attached to the samples.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Smith, A. Using next-generation sequencing to develop a Shigella species threshold and profile faecal samples from suspected diarrhoea cases. Folia Microbiol 66, 399–410 (2021). https://doi.org/10.1007/s12223-020-00846-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-020-00846-w