Abstract
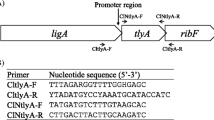
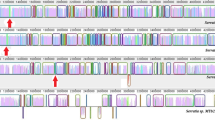
When recombinant plasmid DNA from a genomic DNA library and inverse PCR products of Campylobacter sputorum biovar paraureolyticus LMG17591 strain were analyzed, an approximate 6.5-kb pair region, encoding a urease gene operon, was identified. Within the operon, seven closely spaced and putative open reading frames for ureG, ureH(D), ureA, ureB, ureC, ureE, and ureF were detected in order. A possible overlap was detected between ureG and ureH(D), ureH(D) and ureA, and ureE and ureF. In addition, two putative promoter structures, probable ribosome-binding sites and a putative ρ-independent transcriptional terminator structure were identified. The urease gene operon transcription in the cells was confirmed by the reverse transcription-PCR analysis. A neighbor-joining tree constructed based on the nucleotide sequence information of urease genes showed that C. sputorum biovar paraureolyticus formed a cluster with Arcobacter butzleri, urease-positive thermophilic Campylobacter and some Helicobacter spp., separating those from the other urease-producing bacteria, suggesting a commonly shared ancestry among these organisms.




Similar content being viewed by others
References
Akada JK, Shirai M, Takeuchi H, Tsuda M, Nakazawa T (2000) Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol 36:1071–1084
Beckwith CS, McGee DJ, Mobley HL, Riley LK (2001) Cloning, expression, and catalytic activity of Helicobacter hepaticus urease. Infect Immun 69:5914–5920
Benjamin L (2000) Genes VII. Oxford University Press, Oxford
Bézian MC, Ribou G, Barberis-Giletti C, Mégraud F (1990) Isolation of a urease postive thermophilic variant of Campylobacter lari from a patient with urinary tract infection. Eur J Clin Microbiol Infect Dis 9:895–897
Bolton FJ, Holt AV, Hutchinson DN (1985) Urease-positive thermophilic campylobacters. Lancet I:1217–1218
Ferrero RL, Labigne A (1993) Cloning, expression and sequencing of Helicobacter felis urease genes. Mol Microbiol 9:323–333
Jones BD, Mobley HLT (1989) Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol 171:6414–6422
Kakinuma Y, Iida H, Sekizuka T, Usui K, Murayama O, Takamiya S, Millar BC, Moore JE, Matsuda M (2007) Cloning, sequencing and characterization of a urease gene operon from urease-positive thermophilic Campylobacter (UPTC). J Appl Microbiol 103:252–260
Kakinuma Y, Iida H, Sekizuka T, Taneike I, Takamiya S, Moore JE, Millar BC, Matsuda M (2008) Molecular characterisation of urease genes from urease-positive thermophilic campylobacters (UPTC). Br J Biomed Sci 65:148–152
Matsuda M, Moore JE (2004) Urease-positive thermophilic Campylobacter species. Appl Environ Microbiol 70:4515–4518
Mégraud F, Chevrier D, Desplaces N, Sedallian A, Guesdon JL (1988) Urease-positive thermophilic Campylobacter (Campylobacter laridis variant) isolated from an appendix and from human feces. J Clin Microbiol 26:1050–1051
Mobley HLT, Hausinger RP (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev 53:85–108
Mobley HLT, Island MD, Hausinger RP (1995) Molecular biology of microbial ureases. Microbiol Rev 59:451–480
Mollenhauer-Rektorschek M, Hanauer G, Sachs G, Melchers K (2002) Expression of ureI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res Microbiol 153:659–666
Mulrooney SB, Hausinger RP (1990) Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol 172:5837–5843
Neyrolles O, Ferris S, Behbahani N, Montagnier L, Blanchard A (1996) Organization of Ureaplasma urealyticum urease gene cluster and expression in a suppressor strain of Escherichia coli. J Bacteriol 178:647–655
On SLW, Atabay HI, Corry JEL, Harrington CS, Vandamme P (1998) Emended description of Campylobacter sputorum and revision of its infrasubspecific (biovar) divisions, including C. sputorum biovar paraureolyticus, a urease-producing variant from cattle and humans. Int J Syst Bacteriol 48:195–206
Owen RJ, Costas M, Sloss L, Bolton FJ (1988) Numerical analysis of electrophoretic protein patterns of Campylobacter laridis and allied thermophilic campylobacters from the natural environment. J Appl Bacteriol 65:69–78
Park J-U, Song J-Y, Kwon Y-C, Chung M-J, Jun J-S, Park J-W, Park S-G, Hwang H-R, Choi S-H, Baik S-C, Kang H-L, Youn H-S, Lee W-K, Cho M-J, Rhee K-H (2005) Effect of the urease accessory genes on activation of the Helicobacter pylori urease apoprotein. Mol Cells 20:371–377
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Usui K, Iida H, Ueno H, Sekizuka T, Matsuda M, Murayama O, Millar BC, Moore JE (2006) Genetic heterogeneity of urease gene loci in urease-positive thermophilic Campylobacter (UPTC). Int J Hyg Environ Health 209:541–545
Acknowledgments
This research was partially supported by a research project grant awarded by the Azabu University (Research Services Division) and by Grant-in-Aid for Scientific Research (C; no. 20580346) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to the corresponding author).
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. Kakinuma and K. Hayashi have contributed equally to this study and hence should be considered as equal first authors.
Rights and permissions
About this article
Cite this article
Kakinuma, Y., Hayashi, K., Tazumi, A. et al. Molecular analysis and characterization of a urease gene operon from Campylobacter sputorum biovar paraureolyticus . Folia Microbiol 56, 159–165 (2011). https://doi.org/10.1007/s12223-011-0020-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-011-0020-6