Abstract
Mycobacterium smegmatis, a rapidly growing non-pathogenic mycobacterium, is currently used as a model organism to study mycobacterial genetics. Acetamidase of M. smegmatis is the highly inducible enzyme of Mycobacteria, which utilizes several amide compounds as sole carbon and nitrogen sources. The acetamidase operon has a complex regulatory mechanism, which involves three regulatory proteins, four promoters, and three operator elements. In our previous study, we showed that over-expression of AmiA leads to a negative regulation of acetamidase by blocking the P2 promoter. In this study, we have identified a new positive regulatory protein, AmiC that interacts with AmiA through protein-protein interaction. Gel mobility shift assay showed that AmiC protein inhibits AmiA from binding to the P2 promoter. Interaction of AmiC with cis-acting elements identified its binding ability to multiple regulatory regions of the operon such as P3, OP3, and P1 promoter/operator. Consequently, the addition of inducer acetamide to AmiC complexe trips the complexes, causing AmiC to appear to be the sensory protein for the amides. Homology modeling and molecular docking studies suggest AmiC as a member of Periplasmic binding proteins, which preferentially bind to the inducers and not to the suppressor. Over-expression of AmiC leads to down-regulation of the negative regulator, amiA, and constitutive up-regulation of acetamidase. Based on these findings, we conclude that AmiC positively regulates the acetamidase operon.










Similar content being viewed by others
References
Allen WJ et al (2015) DOCK 6: impact of new features and current docking performance. J Comput Chem 36:1132–1156. https://doi.org/10.1002/jcc.23905
Ames GF (1986) Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem 55:397–425. https://doi.org/10.1146/annurev.bi.55.070186.002145
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Brown AC, Parish T (2006) Instability of the acetamide-inducible expression vector pJAM2 in mycobacterium tuberculosis. Plasmid 55:81–86. https://doi.org/10.1016/j.plasmid.2005.06.005
Chatterjee S, Zhou YN, Roy S, Adhya S (1997) Interaction of gal repressor with inducer and operator: induction of gal transcription from repressor-bound DNA. Proc Natl Acad Sci U S A 94:2957–2962
Daftuar L, Zhu Y, Jacq X, Prives C (2013) Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One 8:e68667. https://doi.org/10.1371/journal.pone.0068667
Draper P (1967) The aliphatic acylamide amidohydrolase of mycobacterium smegmatis: its inducible nature and relation to acyl-transfer to hydroxylamine. J Gen Microbiol 46:111–123
Kim TH, Leslie P, Zhang Y (2014) Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 5:860–871
Komeda H, Kobayashi M, Shimizu S (1996a) Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci U S A 93:4267–4272
Komeda H, Kobayashi M, Shimizu S (1996b) A novel gene cluster including the Rhodococcus rhodochrous J1 nhlBA genes encoding a low molecular mass nitrile hydratase (L-NHase) induced by its reaction product. J Biol Chem 271:15796–15802
Mahenthiralingam E, Draper P, Davis EO, Colston MJ (1993) Cloning and sequencing of the gene which encodes the highly inducible acetamidase of mycobacterium smegmatis. J Gen Microbiol 139:575–583
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Narayanan S, Selvakumar S, Aarati R, Vasan SK, Narayanan PR (2000) Transcriptional analysis of inducible acetamidase gene of mycobacterium smegmatis. FEMS Microbiol Lett 192:263–268
Norman RA, Poh CL, Pearl LH, O'Hara BP, Drew RE (2000) Steric hindrance regulation of the Pseudomonas Aeruginosa amidase operon. J Biol Chem 275:30660–30667. https://doi.org/10.1074/jbc.M000813200
O'Hara BP, Norman RA, Wan PT, Roe SM, Barrett TE, Drew RE, Pearl LH (1999) Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J 18:5175–5186. https://doi.org/10.1093/emboj/18.19.5175
Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ (1997) Regulation of the inducible acetamidase gene of mycobacterium smegmatis. Microbiology 143(Pt 7):2267–2276
Parish T, Turner J, Stoker NG (2001) amiA is a negative regulator of acetamidase expression in mycobacterium smegmatis. BMC Microbiol 1:19
Pearl L, O'Hara B, Drew R, Wilson S (1994) Crystal structure of AmiC: the controller of transcription antitermination in the amidase operon of Pseudomonas Aeruginosa. EMBO J 13:5810–5817
Roberts G, Muttucumaru DG, Parish T (2003) Control of the acetamidase gene of mycobacterium smegmatis by multiple regulators. FEMS Microbiol Lett 221:131–136
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. https://doi.org/10.1006/jmbi.1993.1626
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. https://doi.org/10.1038/nprot.2006.468
Subbian S, Narayanan S (2007) Identification and characterization of the regulatory elements of the inducible acetamidase operon from mycobacterium smegmatis. Can J Microbiol 53:599–606. https://doi.org/10.1139/w06-147
Sundararaman B, Palaniyandi K, Venkatesan A, Narayanan S (2014) Expression, purification and functional characterization of AmiA of acetamidase operon of mycobacterium smegmatis. Microbiol Res 169:873–880. https://doi.org/10.1016/j.micres.2014.02.011
Wilson S, Drew R (1991) Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas Aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol 173:4914–4921
Wilson SA, Wachira SJ, Drew RE, Jones D, Pearl LH (1993) Antitermination of amidase expression in Pseudomonas Aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J 12:3637–3642
Wyborn NR, Mills J, Williams SG, Jones CW (1996) Molecular characterisation of formamidase from Methylophilus methylotrophus. Eur J Biochem 240:314–322
Acknowledgements
Mr. Arunkumar Venkatesan would like to acknowledge the Indian Council for Medical Research (ICMR) for providing a Senior Research Fellowship. We thank Dr. Sameer Hassan and Mr. Yuvaraj for their help in bioinformatics analysis. We would also like to thank Mr. R. Senthilnathan for his technical help.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Fig. S1
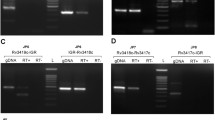
Removal of AmiA from P2 promoter. (A) AmiC removes AmiA from P2 promoter: EMSA was performed for P2 promoter with His-AmiA and AmiC. Lane 1, Labeled P2 promoter alone; Lane 2, addition of His-AmiA alone; lane 3 to 9, AmiC titration with increasing concentration (50, 75, 100, 200, 300, 400 and 500 nM). (GIF 1 kb).
Fig. S2
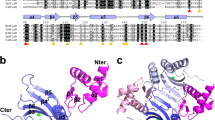
AmiC secondary structure. Secondary structure of AmiC was predicted using PSIPRED server. Rectangle boxes indicate alpha helixes and arrows indicate beta strands. * Represents the periplasmic binding protein domain region. (GIF 883 bytes).
Fig. S3
Graphical representation of probable AmiC binding motif. AmiC DNA binding motif from MEME analyses of P1, P3 and OP3 promoter/operator regions. (GIF 2 kb).
Rights and permissions
About this article
Cite this article
Venkatesan, A., Palaniyandi, K. & Narayanan, S. Molecular characterization of AmiC, a positive regulator in acetamidase operon of Mycobacterium smegmatis . Cell Stress and Chaperones 23, 539–550 (2018). https://doi.org/10.1007/s12192-017-0861-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-017-0861-2