Abstract
Assessing and monitoring the growth and response of bacteria to antibiotics is of crucial importance in research laboratories, as well as in food, environment, medical, and pharmaceutical industrial applications. In this study, Escherichia coli was chosen as the model microorganism to evaluate its response (e.g., growth) to a commercial antibiotic—tetracycline. Thus, the objective of this work was to explore the ability of NIR data combined with machine learning tools (e.g., partial least squares discriminant analysis) to monitor the response and growth of Escherichia coli cultured with different concentrations of tetracycline (ranging from 0 to 50 μg/mL). This study demonstrated a novel method capable of analyzing samples of a complex matrix, while still contained in a 96-well plate. This work will pave the way as a new machine learning method to detect resistance changes in microorganisms without the laborious and, in some cases, time-consuming protocols currently in use in research and by the industry.
Similar content being viewed by others
Introduction
The measurement of the growth phases in bacteria using optical density at 600 nm (OD600) has been the “gold standard” for the industry and research laboratories (Koch and Ehrenfeld 1968; Flickinger and Koch 2010). This method has been extensively used to assess the concentration of cells in a liquid while exhibiting little damage or growth hindrance to cells (Koch and Ehrenfeld 1968). Escherichia coli is the most abundant bacteria that lives predominantly in the intestinal tract of humans and warm-blooded animals (Yoon and Hovde 2008). E. coli has been used as a biological model in a wide range of studies (e.g., basic and applied research) (Tzipori et al. 1985; Jiang et al. 2015). In food, E. coli O157:H7 is a species that can cause acute food poisoning responses in humans, and is occasionally responsible for costly product recalls (Mead and Griffin 1998). E. coli is also used as a fecal contamination indicator species for environmental monitoring, showcasing the ubiquity and therefore importance of the microorganism (Moreno et al. 2008; Elder et al. 2000).
Assessing and monitoring the growth of E. coli is of crucial importance in applied and basic research laboratories, as well as in the food, environment, medical, and pharmaceutical industries (Tzipori et al. 1985; Jiang et al. 2015). Recent rise of antimicrobial resistance (AMR) due to the overuse of antibiotics leads to more resistant strains of bacteria (Tadesse et al. 2012). The rapid adaptability of bacteria to grow and evolve in response to antibiotics has resulted in the need for rapid and high-throughput monitoring tools to quantify and qualify the degree of AMR, and also generate targeted therapeutics for the type of strains analyzed (von Baum and Marre 2005). Development of these tools is urgently needed not only now but for multiple organisms and in a variety of matrices, exacerbated with superbugs and the 2020 COVID-19 pandemic crisis (Murray 2020).
During the last two decades, the so-called process analytical technologies (PAT) have been developed to address real-time monitoring in industrial settings (Hinz 2006). These technologies are based on the concept that quality of a product or process should be evaluated throughout the whole process, by performing real-time measurements during processing instead of carrying out quality control measurements in the final product (Hinz 2006). The PAT guidelines were created concomitantly with developments in process together with the fact that modern process analyzers can provide non-destructive measurements containing information related to biological, physical, and chemical attributes of the materials being processed (Burggraeve et al. 2013). Fermentation monitoring and process control, the implementation of fast analytical tools, such as vibrational spectroscopy, has gained popularity over the last decades. The utilization of vibrational spectroscopy falls into the PAT guidelines as it allows getting real-time information of the process and taking corrective measures (Burggraeve et al. 2013). These techniques (e.g., NIR, MIR, and Raman spectroscopy) seem ideal to analyze broth components and cell culture, because it ensures rapid responses (assay time is usually no longer than 2 min), providing the opportunity to measure several constituents simultaneously (Burggraeve et al. 2013). In recent years, NIR on-line applications have particularly been evaluated with the sample cell connected with the bioreactor through a loop, to evaluate the fermentations of lactic acid production with microaerophilic cultures of Lactobacillus sp. and to monitor the biomass in a fed-batch Escherichia coli process in situ (Burggraeve et al. 2013; Arnold et al. 2002).
Advances and developments in modern analytical instrumentation and spectroscopy have determined an increase in the so-called high-dimensional and high-throughput methods (Cozzolino 2020). Consequently, data generated by both the number of measured variables and the samples can be voluminous. Furthermore, there will be high multiplicities of data (multiple data sources are available) and high frequency during the collection of the data can add to the volumes of data generated. Therefore, the application of these technologies requires the use of data analysis methods and techniques (e.g., machine learning).
The objective of this study was to evaluate the utilization of NIR spectra collected using a 96-well plate combined with machine learning tools to in situ monitor the growth and response of Escherichia coli to increase concentrations of tetracycline (ranging from 0 to 50 μg/mL).
Materials and methods
Samples of Escherichia coli DH5α were cultured overnight in a Luria-Bertani (LB) broth (Sigma-Aldrich, Australia) and subsequently diluted in LB broth to obtain an optical density at 600 nm (OD600). Commonly, LB broth contains 10 g/L peptone, 5 g/L yeast extract, and 10 g/L NaCl. Growth assays were performed in a 96-well plate format, using the CLARIOstar Plus microplate reader (BMG LABTECH Pty. Ltd., 2/24 Carbine Way, Mornington, Victoria, Australia). Details about the instrument specification can be found elsewhere (BMG LABTECH Pty. Ltd., 2/24 Carbine Way, Mornington, Victoria, Australia). Cell growth was monitored over 1-h intervals for 24 h at 37 °C with or without the addition of different levels or concentrations of tetracycline (ranging from 0 to 50 μg/mL). The NIR spectra of the culture samples were automatically recorded using the 96-well microplates (20 μL) in the microplate reader. The microplate reader allows collecting the spectra ranging from 200 to 1000 nm. In this study, the near-infrared range between 750 and 1000 nm (2-nm resolution) was acquired for each sample. Prior to chemometric analysis, the spectra were averaged from 5 repeated spectra. The NIR spectra were recorded in the transmission mode (BMG LABTECH Pty. Ltd., 2/24 Carbine Way, Mornington, Victoria, Australia).
The spectra data were exported from the CLARIOstar Plus microplate reader software in *.csv format to The Unscrambler software (version X, CAMO ASA, Oslo, Norway), where chemometric analysis and spectral pre-processing were carried out. Before any data analysis, pre-processing of the NIR spectra was achieved using the Savitzky-Golay transformation (second derivative, 20-point smoothing, 2nd polynomial order). Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were carried out to identify trends in the spectra as well as to classify the samples. In this study, the samples were the culture (combination) of the microorganisms and the antibiotic treatment, as well as the control (broth), tetracycline (at different concentrations), and the bacteria (bacteria and broth). During PLS-DA, samples were assigned with a dummy number linked with the treatment applied to the samples (e.g., 1 = low (0.1 μg/mL), 2 = medium (25 μg/mL), and 3 = high (50 μg/mL) concentration of tetracycline). Full cross-validation was used to evaluate the models and to minimize the effect of overfitting during model development (PCA and PLS-DA). The root mean square error of cross validation (RMSECV), the coefficient of determination in cross validation (R2), the bias, and the slope were used to evaluate the PLS-DA models developed.
Results and discussion
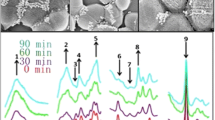
Figure 1 shows the optical density values at 600 nm (OD600) for the different treatments at time zero and at the end of the experiment (24 h). The use of OD600 alone does not provide with enough information about the effect of the addition of antibiotic or any other interactions that might occur during the growth of the bacteria. Thus, the whole scan was analyzed using chemometrics in order to gather more insights about the process. Figure 2 shows the second derivative of the NIR spectra of E. coli samples collected using the 96-well plate. Wavelengths around 764 nm might be associated with O-H overtones (water), around 830 nm with C-H methylene groups, around 846 nm and 874 nm with C-H aromatic groups, and around 910 nm and 928 nm with C-H2 bonds. The wavelength at 970 nm might be associated with O-H groups (water) (Workman Jr. and Weyer 2012; Beć et al. 2020). These wavelengths showed the data-rich NIR spectra collected during the time course of the growth of the bacteria with and without the addition of tetracycline. Principal component analysis was used to interpret the NIR spectra of the E. coli samples collected using the 96-well plate. Figure 3 shows both the PCA score plots where the first two principal components explain 80% of the variability in the data set. Due to the complexity of the data set, it was very difficult to see any clear trends when all samples were analyzed all together. Therefore, PCA models were developed for specific treatments in order to illustrate the ability of the proposed methodology to monitor E. coli growth with the addition of different concentrations of tetracycline. Figure 4 shows the PCA score plot using the highest (50 μg/mL) and lowest concentrations (0 μg/mL) of tetracycline over 24 h of bacteria growth. The PCA score plots demonstrated the capability of NIR combined with multivariate analyses to monitor the changes in E. coli as consequence of the addition of the different concentrations of antibiotic. In both examples, the combination of PC1 and PC2 explained more than 80% of the variation in the data set associated with the addition of antibiotic to the samples.
Principal component score plot for the analysis of E. coli samples analyzed using near-infrared transmission spectroscopy. a Plots for E. coli response to the various concentrations (repeat 1 and repeat 2 are two independent repeats of the same conditions; NB is Luria-Bertani broth made from peptone, yeast extract, sodium chloride). b Plots for E. coli response to the tetracycline concentrations over a period of time (0–24 h)
Principal component score plot for the analysis of E. coli samples analyzed using near-infrared transmission spectroscopy. a The response of E. coli against the high concentration (50 μL/mL) of tetracycline over the course of 24 h. b The response of E. coli against the low concentration (5 μL/mL) of tetracycline over the course of 24 h
Figure 5 (panels a and b) shows the results for the PLS-DA regression models based on the NIR spectra of the samples collected in the microplate reader. The root mean square error of cross validation (RMSECV) and the coefficient of determination (R2) in calibration for the prediction of time were 1.58 h and 0.80, while for the prediction of treatment, these were 2.87 μg/mL and 0.78, respectively. Overall, the PLS-DA models correctly classified 95% of samples belonging to time of growth while only 80% of samples were correctly classified according to treatment (tetracycline concentration). The low classification rates obtained for the prediction of treatment might be explained by the difficulty of the method to identify differences between close concentrations of antibiotic (e.g., between 0.3, 0.4, and 0.5 μg/mL). In Fig. 6, most of the NIR information (loadings) used for the discrimination of samples according to time of growth was contained in wavelengths around 810 nm, 852 nm, 870 nm, 876 nm, 894 nm, 930 nm and 954 nm [18, 19]. Most of these wavelengths were associated with changes in protein, water contents and other chemical compounds present in the bacteria and broth. Some of these wavelengths may be also associated with the high concentration of tetracycline and its effects on bacteria growth (e.g., bacteria are killed by causing changes in the cell membrane) (Chopra and Roberts 2001). It was also observed that the loadings used by the PLS-DA models for the prediction of time show opposite wavelengths (different direction) as the ones observed in the models used to classify samples according to treatment. In this case, the main PLS loadings were observed around 785 nm, 840 nm, 900 nm, and 936 nm associated with changes in protein and dry matter/water content (Workman Jr. and Weyer 2012; Beć et al. 2020). Overall, the discrimination among samples according to time (growth) and treatment (concentration of tetracycline) might be explained by differences in aromatic compounds, lipids and proteins, as well as changes in dry matter (water content).
Partial least squares loadings derived from the calibration models reported in Fig. 4
This study has shown that short wavelengths in the NIR region have the potential to be used as an effective tool to screen E. coli samples on the basis of their spectral information using the 96-well plate. It is well known that the ability of an NIR model to discriminate or identify samples is based on the vibrational responses of specific chemical bonds to NIR radiation. It is highly probable that the higher the variability in these chemical entities, which respond to this range of the electromagnetic spectrum, the better the accuracy of the model developed (Currà et al. 2019). However, the application of NIR to monitor E. coli growth presents challenges related to the complexity of the spectra and the nature of the microbial broths (e.g., changes in the concentration and type of reagents used). The machine learning tools utilized in this study required to transform the raw spectra into useful models and the wide variations in the concentration of a wide range of chemical parameters over the time make the method difficult to understand. Modern microbiological laboratories monitor the bacteria growth rate by measuring the optical density (OD) at a single wavelength over time (Koch and Ehrenfeld 1968; Flickinger and Koch 2010). It is well recognized that the use of OD600 has several advantages, mainly because it is established, and the measurement can be fast and straightforward.
As stated in the “Introduction,” development of these tools is needed in both research and industry. The current antimicrobial resistant crises and the COVID-19 pandemic are demanding rapid and non-invasive tests as that described here. Furthermore, one criterion which is essential with the advent of the so-called superbugs is the need of rapid diagnosis tools that not only monitor growth (OD) but also provide information about the whole system. Although the use of OD600 can be a simple analysis (a single sample analyzed using quartz or plastic cuvettes), the ability of monitoring several samples in a 96-well plate in order to collect measurements over the full growth cycle of a bacteria will be ideal.
Overall, this study has shown the ability of NIR spectroscopy as a safe and rapid method especially when used to analyze the addition of antibiotics during bacteria growth. This experiment setup has also shown that complex spectra can be used to monitor the interactions between the bacteria and perturbations in the system where the bacteria are cultured such as media (e.g., addition of antibiotic) or time. The potential to extract large amounts of data from a single multi-wavelength measurement makes NIR spectroscopy a powerful characterization tool for in-lab or industrial applications.
Conclusions
In this study, it was demonstrated that machine learning (e.g., partial least squares discriminant analysis) methods in combination with NIR data can be used as a rapid classification tool to monitor resistant and non-resistant strains treated with different concentrations of tetracycline. This novel application also demonstrated that it is possible to monitor and differentiate treatments. The PCA has provided with a simple visual tool to recognize changes in the bacteria (e.g., growth), having additional advantages in comparison with the use of OD600 alone. The interpretation of the PCA and PLS loadings has provided with additional information related with composition of the samples. Due to the unique experimental setup explored, the same test could be used for obtaining practical information on the type, level of resistance, and dose of antibiotic necessary to establish an optimum diagnosis, treatment, and decontamination strategies.
The data has also shown that adding different concentrations of broad-spectrum antibiotics to growth medium creates unique changes in the NIR spectra. However, one of the limitations of this type of experimental protocols might be related with the number of antibiotics that can be used in a 96-well plate, and the number of replicates. Further validation with the addition of more samples and different types and combinations of antibiotics must be evaluated before the methods are fully implemented in routine analysis.
References
Arnold SA, Gaensakoo R, Harvey LM, McNeil B (2002) Use of at-line and in-situ near-infrared spectroscopy to monitor biomass in an industrial fed-batch Escherichia coli process. Biotechnol Bioeng 80:405–413
Beć KB, Grabska JE, Huck CW (2020) Near-infrared spectroscopy in bio-applications. Molecules 25:2948. https://doi.org/10.3390/molecules25122948
Burggraeve A, Monteyne T, Vervaet C, Remon JP, Beer TD (2013) Process analytical tools for monitoring, understanding, and control of pharmaceutical fluidized bed granulation: a review. Eur J Pharm Biopharm 83:2–15
Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260
Cozzolino D (2020) The sample, the spectra and the maths-the critical pillars in the development of robust and sound applications of vibrational spectroscopy. Molecules 25:3674
Currà A, Gasbarrone R, Cardillo A, Trompetto C, Fattapposta F, Pierelli F, Missori P, Bonifazi G, Serranti S (2019) Near-infrared spectroscopy as a tool for in vivo analysis of human muscles. Sci Rep 9:8623
Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW (2000) Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci 97:2999–3003
Flickinger MC, Koch AL (2010) Microbial growth measurement. In: Flickinger MC (ed) Encyclopedia of industrial biotechnology, pp 1–11
Hinz DC (2006) Process analytical technologies in the pharmaceutical industry: the FDA’s PAT initiative. Anal Bioanal Chem 384:1036–1042
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514
Koch AL, Ehrenfeld E (1968) The size and shape of bacteria by light scattering measurements. Biochim Biophys Acta Gen Subj 165:262–273
Mead PS, Griffin PM (1998) Escherichia coli O157:H7. Lancet 352:1207–1212
Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G (2008) Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol 46:2529–2534
Murray AK (2020) The novel coronavirus COVID-19 outbreak: global implications for antimicrobial resistance. Front Microbiol 11:1020–1020
Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF (2012) Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg Infect Dis 18:741–749
Tzipori S, Robins-Browne RM, Gonis G, Hayes J, Withers M, McCartney E (1985) Enteropathogenic Escherichia coli enteritis: evaluation of the gnotobiotic piglet as a model of human infection. Gut 26:570–578
von Baum H, Marre R (2005) Antimicrobial resistance of Escherichia coli and therapeutic implications. International journal of medical microbiology : IJMM 295:503–511
Workman J Jr, Weyer L (2012) Practical guide to interpretive near-infrared spectroscopy, 2nd edn. Taylor & Francis Group, CRC Press, Boca Raton, London, New York
Yoon JW, Hovde CJ (2008) All blood, no stool: enterohemorrhagic Escherichia coli O157:H7 infection. J Vet Sci 9:219–231
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent not applicable.
Conflict of interest
Vi Khanh Truong declares that he has no conflict of interest. James Chapman declares that he has no conflict of interest. Daniel Cozzolino declares that he has no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Truong, V.K., Chapman, J. & Cozzolino, D. Monitoring the Bacterial Response to Antibiotic and Time Growth Using Near-infrared Spectroscopy Combined with Machine Learning. Food Anal. Methods 14, 1394–1401 (2021). https://doi.org/10.1007/s12161-021-01994-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-021-01994-6