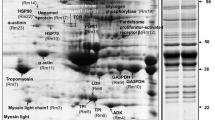
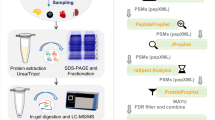
Abstract
Sea bass represents one of the main fish products in the market. Most of it comes from farming and is bred in different conditons with respect to the wild fish. Differences may thus be expected. In this study, a proteomic profile of farmed and wild sea bass samples was performed, employing a fractionation strategy where peptide samples were first separated by 2D chromatography. The peptides were finally analyzed by shotgun proteomics workflow combined to tandem MS. The chosen fractionation approach was successful allowing to greatly improve the fish muscle protein characterization and detect some interesting differences between wild fish and farmed sea bass. Sixty-nine proteins were overexpressed in farmed fish samples, whereas 182 proteins were underexpressed. Some of these proteins could be related to the breeding conditions and the diet with which fishes were fed, thus providing some interesting results for assessing food quality based on a comprehensive proteomic study.





Similar content being viewed by others
References
Addis MF (2013) Farmed and wild fish. In: Toldrá F, Nollet LML (eds) Proteomics in foods. Springer US, Boston, pp 181–203
Addis MF, Cappuccinelli R, Tedde V et al (2010) Proteomic analysis of muscle tissue from gilthead sea bream (Sparus aurata, L.) farmed in offshore floating cages. Aquaculture 309:245–252. doi:10.1016/j.aquaculture.2010.08.022
Alami-Durante H, Cluzeaud M, Duval C et al (2014) Early decrease in dietary protein:energy ratio by fat addition and ontogenetic changes in muscle growth mechanisms of rainbow trout: short- and long-term effects. Br J Nutr 112:674–687. doi:10.1017/S0007114514001391
Alasalvar C, Taylor KD, Zubcov E et al (2002) Differentiation of cultured and wild sea bass (Dicentrarchus labrax): total lipid content, fatty acid and trace mineral composition. Food Chem 79:145–150. doi:10.1016/S0308-8146(02)00122-X
Bhatia VN, Perlman DH, Costello CE, McComb ME (2009) Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem 81:9819–9823. doi:10.1021/ac901335x
Bosworth CA, Chou C-W, Cole RB, Rees BB (2005) Protein expression patterns in zebrafish skeletal muscle: initial characterization and the effects of hypoxic exposure. Proteomics 5:1362–1371. doi:10.1002/pmic.200401002
Brownridge P, de Mello LV, Peters M et al (2009) Regional variation in parvalbumin isoform expression correlates with muscle performance in common carp (Cyprinus carpio). J Exp Biol 212:184–193. doi:10.1242/jeb.021857
Capriotti AL, Caruso G, Cavaliere C et al (2013a) Proteome investigation of the non-model plant pomegranate (Punica granatum L.) Anal Bioanal Chem 405:9301–9309. doi:10.1007/s00216-013-7382-3
Capriotti AL, Cavaliere C, Foglia P et al (2013b) Proteomic platform for the identification of proteins in olive (Olea europaea) pulp. Anal Chim Acta 800:36–42. doi:10.1016/j.aca.2013.09.014
Capriotti AL, Cavaliere C, Piovesana S et al (2016) Recent trends in the analysis of bioactive peptides in milk and dairy products. Anal Bioanal Chem:1–9. doi:10.1007/s00216-016-9303-8
Costa R, Albergamo A, Piparo M et al (2017) Multidimensional gas chromatographic techniques applied to the analysis of lipids from wild-caught and farmed marine species. Eur J Lipid Sci Technol 119:1600043. doi:10.1002/ejlt.201600043
Cox J, Hein MY, Luber CA, Paron I (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. doi:10.1074/mcp.M113.031591
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi:10.1038/nbt.1511
D’Attoma A, Grivel C, Heinisch S (2012) On-line comprehensive two-dimensional separations of charged compounds using reversed-phase high performance liquid chromatography and hydrophilic interaction chromatography. Part I: Orthogonality and practical peak capacity considerations. J Chromatogr A 1262:148–159. doi:10.1016/j.chroma.2012.09.028
Deeb SJ, D’Souza RCJ, Cox J et al (2012) Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics 11:77–89. doi:10.1074/mcp.M111.015362
Diez A, Menoyo D, Pe S et al (2007) Conjugated linoleic acid affects lipid composition , metabolism , and gene expression in gilthead sea bream ( Sparus aurata L ) 1–3. J Nutr 137:1363–1369
Domon B (2006) Mass spectrometry and protein analysis. Science 312:212–217. doi:10.1126/science.1124619
Fercha A, Capriotti AL, Caruso G et al (2013) Gel-free proteomics reveal potential biomarkers of priming-induced salt tolerance in durum wheat. J Proteome 91:486–499. doi:10.1016/j.jprot.2013.08.010
Focant B, Vandewalle P, Huriaux F (2003) Expression of myofibrillar proteins and parvalbumin isoforms during the development of a flatfish, the common sole Solea solea: comparison with the turbot Scophthalmus maximus. Comp Biochem Physiol B Biochem Mol Biol 135:493–502. doi:10.1016/S1096-4959(03)00116-7
FAO (2016) The state of world Fisheries and Aquaculture 2016. Contributing to food security andnutrition for all. Rome, p 200
Forné I, Abián J, Cerdà J (2010) Fish proteome analysis: model organisms and non-sequenced species. Proteomics 10:858–872. doi:10.1002/pmic.200900609
Gebriel M, Uleberg K-E, Larssen E et al (2010) Cod (Gadus morhua) muscle proteome cataloging using 1D-PAGE protein separation, nano-liquid chromatography peptide fractionation, and linear trap quadrupole (LTQ) mass spectrometry. J Agric Food Chem 58:12307–12312. doi:10.1021/jf103009r
Griesmeier U, Vázquez-Cortès S, Bublin M et al (2010) Expression levels of parvalbumins determine allergenicity of fish species. Allergy 65:191–198. doi:10.1111/j.1398-9995.2009.02162.x
Ghaedi G, Keyvanshokooh S, Akhlaghi M (2016) Proteomic analysis of muscle tissue from rainbow trout (Oncorhynchus mykiss) fed dietary β-glucan. Iran J Vet Res 17:184–189
Hamelin M, Sayd T, Chambon C et al (2006) Proteomic analysis of ovine muscle hypertrophy. J Anim Sci 84:3266. doi:10.2527/jas.2006-162
Kikuchi K, Yamashita M, Watabe S, Aida K (1995) The warm temperature acclimation-related 65-kDa protein, Wap65, in goldfish and its gene expression. J Biol Chem 270:17087–17092. doi:10.1074/jbc.270.29.17087
Kuehn A, Swoboda I, Arumugam K et al (2014) Fish allergens at a glance: variable allergenicity of parvalbumins, the major fish allergens. Front Immunol. doi:10.3389/fimmu.2014.00179
Monti G, De Napoli L, Mainolfi P et al (2005) Monitoring food quality by microfluidic electrophoresis, gas chromatography, and mass spectrometry techniques: effects of aquaculture on the sea bass ( Dicentrarchus labrax). Anal Chem 77:2587–2594. doi:10.1021/ac048337x
Morais S, Silva T, Cordeiro O et al (2012) Effects of genotype and dietary fish oil replacement with vegetable oil on the intestinal transcriptome and proteome of Atlantic salmon (Salmo salar). BMC Genomics 13:448. doi:10.1186/1471-2164-13-448
Morzel M, Chambon C, Lefèvre F et al (2006) Modifications of trout ( Oncorhynchus mykiss ) muscle proteins by preslaughter activity. J Agric Food Chem 54:2997–3001. doi:10.1021/jf0528759
Piovesana S, Capriotti AL, Caruso G et al (2016a) Labeling and label free shotgun proteomics approaches to characterize muscle tissue from farmed and wild gilthead sea bream (Sparus aurata). J Chromatogr A 1428:193–201. doi:10.1016/j.chroma.2015.07.049
Piovesana S, Capriotti AL, Cavaliere C et al (2016b) New magnetic graphitized carbon black TiO 2 composite for phosphopeptide selective enrichment in shotgun phosphoproteomics. Anal Chem 88:12043–12050. doi:10.1021/acs.analchem.6b02345
Reddish JM, St-Pierre N, Nichols A et al (2008) Proteomic analysis of proteins associated with body mass and length in yellow perch, Perca flavescens. Proteomics 8:2333–2343. doi:10.1002/pmic.200700533
Sauer S, Luge T (2015) Nutriproteomics: facts, concepts, and perspectives. Proteomics 15:997–1013. doi:10.1002/pmic.201400383
Stein DR, Hu X, Mccorrister SJ et al (2013) High pH reversed-phase chromatography as a superior fractionation scheme compared to off-gel isoelectric focusing for complex proteome analysis. Proteomics 13:2956–2966. doi:10.1002/pmic.201300079
Tacchi L, Secombes CJ, Bickerdike R et al (2012) Transcriptomic and physiological responses to fishmeal substitution with plant proteins in formulated feed in farmed Atlantic salmon (Salmo salar). BMC Genomics 13:363. doi:10.1186/1471-2164-13-363
Tahmasebi-Kohyani A, Keyvanshokooh S, Nematollahi A et al (2012) Effects of dietary nucleotides supplementation on rainbow trout (Oncorhynchus mykiss) performance and acute stress response. Fish Physiol Biochem 38:431–440. doi:10.1007/s10695-011-9524-x
Tedesco S, Mullen W, Cristobal S (2014) High-throughput proteomics: a new tool for quality and safety in fishery products. Curr Protein Pept Sci 15:118–133. doi:10.2174/1389203715666140221120219
Terova G, Pisanu S, Roggio T et al (2014) Proteomic profiling of sea bass muscle by two-dimensional gel electrophoresis and tandem mass spectrometry. Fish Physiol Biochem 40:311–322. doi:10.1007/s10695-013-9855-x
Vizcaíno JA, Csordas A, Del-Toro N et al (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi:10.1093/nar/gkv1145
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This research was supported by the framework of the Research Project “PRIN 2012”: assessment of quality and safety of Mediterranean seafoods by “omics” sciences, supported by the Italian Ministry of University and Scientific Research, no. 2012TLC44W.
Conflict of Interest
Riccardo Zenezini Chiozzi declares that he has no conflict of interest. Anna Laura Capriotti declares that she has no conflict of interest. Chiara Cavaliere declares that she has no conflict of interest. Giorgia La Barbera declares that she has no conflict of interest. Carmela Maria Montone declares that she has no conflict of interest. Susy Piovesana declares that she has no conflict of interest. Aldo Laganà declares that he has no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed Consent
Not applicable.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chiozzi, R.Z., Capriotti, A.L., Cavaliere, C. et al. Label-Free Shotgun Proteomics Approach to Characterize Muscle Tissue from Farmed and Wild European Sea Bass (Dicentrarchus labrax). Food Anal. Methods 11, 292–301 (2018). https://doi.org/10.1007/s12161-017-0999-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-017-0999-7