Abstract
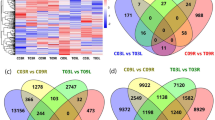
The salt-tolerant capability of the candidate bioenergy crop prairie cordgrass greatly surpasses that of previously characterized prairie grass species and most other plants. To understand the mechanism of inherited salt tolerance, we compared phenotypic and genetic qualities in half-sib families of prairie cordgrass after salt treatment. Each family was treated with a 400 mM NaCl solution or a water control and then measured for various health phenotypes. Phenotypes associated with salt tolerance were shown to be moderately heritable between parent and offspring. RNA-seq analysis revealed differential regulation in unique pathways including metabolism, signaling, photosynthesis, and the circadian rhythm. The studies herein suggest that alternative regulation of the photosynthetic pathway could confer increased salt resistance in halophytes and can be monitored phenotypically or genetically in breeding programs. The improvement of salt-tolerant traits in prairie cordgrass would increase its potential to be grown as a bioenergy crop on lands that are not suitable for the growth of food crops.



Similar content being viewed by others
References
Hitchcock AS from OC, [from old catalog] A (1935) Manual of the grasses of the United States
Boe A, Arvid B, Lee DK (2007) Genetic variation for biomass production in prairie cordgrass and switchgrass. Crop Sci 47:929
Guo J, Jia G, Santanu T et al (2015) Phenotypic and biomass yield variations in natural populations of prairie cordgrass (Spartina pectinata Link) in the USA. Bioenergy Res 8:1371–1383
Boe A, Owens V et al (2009) Morphology and biomass production of prairie cordgrass on marginal lands. GCB Bioenergy 1:240–250
Kim S, Sumin K, Lane Rayburn A et al (2011) Salinity effects on germination and plant growth of prairie cordgrass and switchgrass. Bioenergy Res 5:225–235
Jones MB, John F, Hodkinson TR (2014) Morphological and physiological traits for higher biomass production in perennial rhizomatous grasses grown on marginal land. GCB Bioenergy 7:375–385
Helms D (1961) Land capability classification. US Department of Agriculture, Washington D.C
Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Knight H, Heather K, Trewavas AJ, Knight MR (1997) Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J 12:1067–1078
Guo Y (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell Online 13:1383–1400
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci U S A 97:3735–3740
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A 97:6896–6901
Munnik T, Ligterink W, Meskiene II et al (1999) Distinct osmo-sensing protein kinase pathways are involved in signaling moderate and severe hyper-osmotic stress. Plant J 20:381–388
Mikolajczyk M (2000) Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell Online 12:165–178
Hoyos ME (2000) Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol 122:1355–1364
Mizoguchi T, Ichimura K, Yoshida R, Shinozaki K (2000) MAP kinase cascades in Arabidopsis: their roles in stress and hormone responses. Results Probl Cell Differ 27:29–38
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG et al (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104–106
Kasuga M, Liu Q, Miura S et al (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340
Xiong L, Liming X, Manabu I et al (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13:2063
Iuchi S, Kobayashi M, Taji T et al (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333
Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci U S A 96:15354–15361
Thompson AJ, Jackson AC, Symonds RC et al (2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23:363–374
Liotenberg S, Sylviane L, Helen N, Annie M-P (1999) Molecular biology and regulation of abscisic acid biosynthesis in plants. Plant Physiol Biochem 37:341–350
Seo M, Peeters AJ, Koiwai H et al (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci U S A 97:12908–12913
Walia H, Wilson C, Condamine P et al (2007) Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 30:410–421
Gedye K, Gonzalez-Hernandez JL et al (2010) Investigation of the transcriptome of prairie cord grass, a new cellulosic biomass crop. Plant Genome J 3:69
Nah G, Lee M, Kim D-S et al (2016) Transcriptome analysis of Spartina pectinata in response to freezing stress. PLoS One 11:e0152294
Nyquist WE, Baker RJ (1991) Estimation of heritability and prediction of selection response in plant populations. CRC Crit Rev Plant Sci 10:235–322
Core Team R (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
de Mendiburu F, Simon R (2015) Agricolae—ten years of an open source statistical tool for experiments in breeding, agriculture and biology. Peer J Preprints 3:e1404v1
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Stark C (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D535–D539
Thimm O, Bläsing O, Gibon Y et al (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Reitz LP, Salmon SC (1968) Origin, history, and use of Norin 10 wheat. Crop Sci 8:686
Jain HK, Kharkwal MC (2012) Plant breeding: Mendelian to molecular approaches. Springer, Netherlands
Hotta H, Aoki N, Matsuda T, Adachi T (1998) Molecular analysis of a novel protein kinase in maturing rice seed. Gene 213:47–54
Boudsocq M, Barbier-Brygoo H, Laurière C (2004) Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 279:41758–41766
Friedberg F, Felix F (1988) Calcium binding protein families: the “E–F hand” family. Biochem Educ 16:35–36
Chen C, Sun X, Duanmu H et al (2015) GsCML27, a gene encoding a calcium-binding Ef-hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses. PLoS One 10:e0141888
Lockhart J (2013) Salt of the earth: ethylene promotes salt tolerance by enhancing Na/K homeostasis. Plant Cell 25:3150
Jiang C, Belfield EJ, Cao Y et al (2013) An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25:3535–3552
Kerstiens G, Gerhard K, Wlodek T et al (2002) Sodium-related partial stomatal closure and salt tolerance of Aster tripolium. New Phytol 153:509–515
Robertson RN, Wilkins M (1948) Quantitative relation between salt accumulation and salt respiration in plant cells. Nature 161:101
Hennessey TL, Field CB (1991) Circadian rhythms in photosynthesis : oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol 96:831–836
Dodd AN, Salathia N, Hall A et al (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309:630–633
Sanchez A, Alfredo S, Jieun S, Davis SJ (2011) Abiotic stress and the plant circadian clock. Plant Signal Behav 6:223–231
Grundy J, Stoker C, Carré IA (2015) Circadian regulation of abiotic stress tolerance in plants. Front Plant Sci 6:648
Lai AG, Doherty CJ, Mueller-Roeber B et al (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl AcadSci U S A 109:17129–17134
Nakamichi N, Kusano M, Fukushima A et al (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50:447–462
Nakamichi N, Kiba T, Henriques R et al (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605
SX L, Knowles SM, Andronis C et al (2009) CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol 150:834–843
Xie Q, Wang P, Liu X et al (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26:2843–2857
Xing H, Wang P, Cui X et al (2015) LNK1 and LNK2 recruitment to the evening element require morning expressed circadian related MYB-like transcription factors. Plant Signal Behav 10:e1010888
Yanhui C, Xiaoyuan Y, Kun H et al (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124
Ledger S, Strayer C, Ashton F et al (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26:15–22
Preston JC, Jorgensen SA, Jha SG (2014) Functional characterization of duplicated suppressor of overexpression of Constans 1-like genes in petunia. PLoS One 9:e96108
Drabe ova J, Drabe ova J, Chab D et al (2014) A dark-light transition triggers expression of the floral promoter CrFTL1 and downregulates CONSTANS-like genes in a short-day plant Chenopodium rubrum. J Exp Bot 65:2137–2146
Hassidim M, Harir Y, Yakir E et al (2009) Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230:481–491
Min J-H, Chung J-S, Lee K-H, Kim CS (2015) The CONSTANS-like 4 transcriptionfactor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J Integr Plant Biol 57:313–324
Acknowledgements
This study was funded by a grant of the NC Sun Grant Initiative to A. Boe and J.L. Gonzalez-Hernandez. M. Robben and E. Thibault were supported by a REU grant from the National Science Foundation (DBI-1263335). A. Boe, Y. Wu and J.L. Gonzalez-Hernandez acknowledge support from the South Dakota Agricultural Experimental Station.
Author information
Authors and Affiliations
Contributions
Experiments were designed by J.G.H., A.B, and Y.W. Salt treatments, plant cultivation, and sample collection were performed in part by M.R., E.T., Y.W., and A.B. RNA was extracted by M.R., E.T., and M.T. RNA-seq analysis and data interpretation was done by M.R. and J.G. H. First draft of the manuscript was prepared by M.R.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Robben, M., Thibault, E., Tran, M. et al. Transcriptome Analysis of the Heritable Salt Tolerance of Prairie Cordgrass (Spartina pectinata Link). Bioenerg. Res. 11, 106–114 (2018). https://doi.org/10.1007/s12155-017-9881-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-017-9881-0