Abstract
Objective
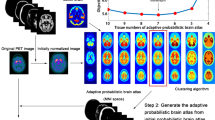
Digital brain template and atlas designed for a specific group provide advantages for the analysis and interpretation of neuroimaging data, but require a significant workload for development. We developed a simple method to create customized brain atlas for diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL) tool using FreeSurfer-generated volume-of-interest (FSVOI) images and validated.
Methods
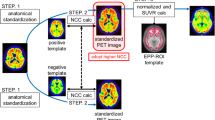
18F-florbetaben positron emission tomography (PET) and magnetic resonance (MR) imaging data were obtained from 248 participants of Alzheimer’s disease spectrum (from cognitively normal to Alzheimer’s disease dementia). To create a customized atlas, MR images of 84 amyloid-negative controls were first processed with FreeSurfer to obtain individual FSVOI and with DARTEL tool to create DARTEL template. Individual FSVOI images were spatially normalized, and each voxel was then labelled with a VOI label with maximum probability. Using these template and atlas, all images were normalized, and the regional standardized uptake value ratios (SUVR) were measured.
Results
18F-florbetaben SUVR values measured with customized atlas showed excellent one-to-one correlation with SUVR measured with individual FSVOI in all regions, and thereby showed almost identical between-group comparison results and outperformed the classic methods.
Conclusions
Customized FreeSurfer-based brain atlas for DARTEL tool is easy to create and useful for the analysis of PET and MR images with high adaptability and reliability for broad research purposes.



Similar content being viewed by others
References
Cabezas M, Oliver A, Llado X, Freixenet J, Cuadra MB. A review of atlas-based segmentation for magnetic resonance brain images. Comput Methods Progr Biomed. 2011;104:e158–e177177.
Evans AC, Janke AL, Collins DL, Baillet S. Brain templates and atlases. Neuroimage. 2012;62:911–22.
Mandal PK, Mahajan R, Dinov ID. Structural brain atlases: design, rationale, and applications in normal and pathological cohorts. J Alzheimers Dis. 2012;31(Suppl 3):S169–S188188.
Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–55.
Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22.
Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80.
Su Y, D'Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS ONE. 2013;8:e73377.
Greve DN, Svarer C, Fisher PM, Feng L, Hansen AE, Baare W, et al. Cortical surface-based analysis reduces bias and variance in kinetic modeling of brain PET data. Neuroimage. 2014;92:225–36.
Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–66.
Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113.
Matsuda H, Mizumura S, Nemoto K, Yamashita F, Imabayashi E, Sato N, et al. Automatic voxel-based morphometry of structural MRI by SPM8 plus diffeomorphic anatomic registration through exponentiated lie algebra improves the diagnosis of probable Alzheimer Disease. AJNR Am J Neuroradiol. 2012;33:1109–14.
Takahashi R, Ishii K, Miyamoto N, Yoshikawa T, Shimada K, Ohkawa S, et al. Measurement of gray and white matter atrophy in dementia with Lewy bodies using diffeomorphic anatomic registration through exponentiated lie algebra: A comparison with conventional voxel-based morphometry. AJNR Am J Neuroradiol. 2010;31:1873–8.
Martino ME, de Villoria JG, Lacalle-Aurioles M, Olazaran J, Cruz I, Navarro E, et al. Comparison of different methods of spatial normalization of FDG-PET brain images in the voxel-wise analysis of MCI patients and controls. Ann Nucl Med. 2013;27:600–9.
Villemagne VL, Mulligan RS, Pejoska S, Ong K, Jones G, O'Keefe G, et al. Comparison of 11C-PiB and 18F-florbetaben for Abeta imaging in ageing and Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2012;39:983–9.
Kang Y, Na DL. Seoul Neuropsychological screening battery (SNSB). Incheon, South Korea: Human Brain Research & Consulting Co.; 2003.
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–44.
Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–8.
Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: phase 3 study. Alzheimers Dement. 2015;11:964–74.
Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–51.
Funding
This research was supported by a grant from Korean Neurological Association (KNA-19-MI-12), faculty research Grant of Yonsei University College of Medicine for (6-2018-0068), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2017R1A2B2006694) and the Ministry of Education (NRF-2018R1D1A1B07049386), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI18C1159).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baek, M., Cho, H., Ryu, Y. et al. Customized FreeSurfer-based brain atlas for diffeomorphic anatomical registration through exponentiated lie algebra tool. Ann Nucl Med 34, 280–288 (2020). https://doi.org/10.1007/s12149-020-01445-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-020-01445-y