Abstract
Most previous studies of homegardens have used labor-intensive boots-on-the-ground plant surveys, owner questionnaires, and interviews, limiting them to at most a few hundred homegardens. We show that automated analysis of publicly available imagery can enable surveys of much greater scale that can augment these traditional data sources. Specifically, we demonstrate the feasibility of using the high-resolution street-level photographs in Google Street View and an object-detection network (RetinaNet) to create a large-scale high-resolution survey of the prevalence of at least six plant species widely grown in road-facing homegardens in Thailand. Our research team examined 4000 images facing perpendicular to the street and located within 10 m of a homestead, and manually outlined all perceived instances of eleven common plant species. A neural network trained on these tagged images was used to detect instances of these species in approximately 150,000 images constituting views of roughly one in every ten homesteads in five provinces of northern Thailand. The results for six of the plant species were visualized as heatmaps of both the average number of target species detected in each image and individual species prevalence, with spatial averaging performed at scales of 500 m and 2.5 km. Urban-rural contrasts in the average number of target species in each image are quantified, and large variations are observed even among neighboring villages. Spatial heterogeneity is seen to be more pronounced for banana and coconut than for other species. Star gooseberry and papaya are more frequently present immediately outside of towns while dracaena and mango persist into the cores of towns.
Similar content being viewed by others
Introduction
The focus of this paper is homegardens, which have historically provided families with a location near the homestead to grow a variety of plants important to their well-being (Idohou et al. 2014). Fruits, vegetables, and medicinal plants are grown for home consumption, and plants for ornament, shade, and other benefits, are also grown and maintained by family labor (Cruz Garcia and Struik 2015; Lattirasuvan et al. 2010). Typically comprising diverse species in multiple structures with multiple functions, homegardens are considered to be complex integrated agricultural ecosystems (Das and Das 2005; Fernandes and Nair 1986; Kumar and Nair 2004) and traditional conservation systems (Galluzzi et al. 2010). Due to their multifaceted importance and complexity, researchers have conducted studies to understand homegardens’ multiple benefits, including their potential to maintain and enhance biodiversity (Clarke et al. 2014; Galluzzi et al. 2010; Trinh et al. 2003), food security (Berti et al. 2004; Schreinemachers et al. 2015), resilience (Colding and Barthel 2013), and sustainable development (Weinberger 2013).
Most studies of homegardens have focused on compiling comprehensive lists of species and quantifying species richness in a few dozen to a few hundred gardens, using boots-on-the-ground plant surveys and owner questionnaires and interviews (Cruz Garcia and Struik 2015; George and Christopher 2019; Jemal et al. 2018; Kabir and Webb 2008; Mathewos et al. 2018; Pala et al. 2019; Panyadee et al. 2018; Panyadee et al. 2019; Rayol et al. 2019; Serrano-Ysunza et al. 2018; Tadesse et al. 2019; Vibhuti et al. 2018; Webb and Kabir 2009; Whitney et al. 2018; Woldeamanual et al. 2018; Yamane et al. 2018). Sampling methods are frequently random (George and Christopher 2019; Jemal et al. 2018; Mathewos et al. 2018; Pala et al. 2019; Tadesse et al. 2019; Yamane et al. 2018) and convenience sampling has also been used in these homegarden studies (Panyadee et al. 2018; Panyadee et al. 2019). Some studies focus in particular on woody plants because they are stable across longer periods of time and produce large amounts of useful fruits and leaves (Panyadee et al. 2016). Many studies of woody plants in homegardens conduct field inventories to count individual occurrences (Jemal et al. 2018; Kabir and Webb 2008; Molla and Kewessa 2015; Panyadee et al. 2016; Tadesse et al. 2019), or conduct focused inventories within designated study plots within the gardens (Pala et al. 2019; Shumi et al. 2018).
In this paper we build on our previous work (Ringland et al. 2019), to apply a technique that capitalizes on the availability of an enormous collection of street-level images along road networks - in Google Street View (GSV) - to conduct an automated survey of a selected set of common species grown in Thai homegardens. Our goal was to test if we can use GSV and an object-detection network to accurately identify and count instances of a significant number of commonly grown plant species in Thai homegardens over a large geographic region. We conducted this study in the hope that the large quantity of data that could be produced by our approach could be combined with that from detailed research on the ground to inform attempts to understand multiple aspects of homegardening practices, including: the significant variation in garden composition across regions (Coomes and Ban 2004) and by season (Cruz Garcia and Struik 2015); the factors that influence the selection of particular species, such as the modes of utilization of specific plants (Gajaseni and Gajaseni 1999) and geographical location (Huai et al. 2011); and cultural backgrounds and personal preferences (Srithi et al. 2012).
The computer vision tool we have developed to collect and analyze the GSV images is also much faster than related alternatives such as car surveys or manual GSV image analysis (Berland and Lange 2017; Deus et al. 2016). This has allowed us to create a survey capturing almost one million individual instances of some of the most commonly found plant species in road-facing homegardens in Thailand. As compared to field inventories, we expect that our approach can save time and money for homegarden research. This proof-of-concept study is the first of its kind; this article provides information on capabilities and limitations of this approach, and affirms the potential of automated GSV image interpretation in homegarden research.
Methods
Study area
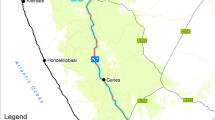
The study area [Fig. 1] comprises five contiguous provinces of northern Thailand: Chiang Mai, Lamphun, Lampang, Phayao, and Phrae (red-tinted region in the figure), with a total area of just over 50,000 km2 (roughly 270 km by 320 km, from longitude 98.0° to 100.6°E, latitude 17.2° to 20.1°N), and a combined population of just under 3.7 million as of 2011 (National Statistical Office 2011). Topographically, this area consists largely of plains where most inhabitants live separated by sparsely inhabited mountain ranges which mostly run roughly north-south. In the 17 provinces making up Thailand’s northern region, 90.2% of the population lives in detached housing, and 56% of the economically active population over age 15 works in skilled agriculture and fisheries. 74% of the population over 10 years old engaged in some kind of agricultural activity on their own lands (National Statistical Office 2011). Climatically, northern Thailand is warm or hot year-round with daily highs rarely lower than 27 °C, and daily lows usually above 18 °C except December to February. April is the hottest month, when temperatures can reach 37 °C. There is a nine-month rainy period from early March to early December with peaks in late May and late August. Skies are predominantly overcast from mid-April to mid-October, and often clear or partly so during the rest of the year (Cedar Lake Ventures 2020) It can be easily verified by dropping into GSV at random locations that almost every dwelling in this region has a garden adjacent to it, frequently containing the species detected in this study.
The study area in northern Thailand, tinted orange in this map, comprises the provinces of Chiang Mai, Lampang, Lamphun, Phayao, and Phrae. Inset at upper left: the location of Thailand and the study area in Southeast Asia. Use of main base map CC BY-SA 3.0 (Damm 2008). Use of inset base map CC BY-SA 3.0 (Zuanzuanfuwa 2012)
Panorama discovery and image acquisition
Since there is no public master list of available GSV imagery, the task of finding all available panoramas in the region of interest was accomplished by probing the GSV metadata application programming interface (API), using URLs of the form https://maps.googleapis.com/maps/api/streetview/metadata?location=14.54173,104.74517&key = API_KEY_HERE, which requests a panorama near latitude 14.54173°N, longitude 104.74517°E. An API key can be obtained without charge from Google. If a panorama exists close to the specified latitude and longitude, this request returns the panorama’s precise location, its month and year of capture, and its unique “panoid” which can subsequently be used to obtain imagery.
We performed panorama discovery by district (amphoe) within the five provinces. In each district, starting with one or more randomly selected “seed” panoramas, we probed for panoramas near previously discovered ones until the vicinity of every discovered panorama had been thoroughly probed. This activity is represented schematically as step 1 in Fig. 2, which is a flow chart of our entire process. The effective road-speed of panorama discovery was 60–80 km/h per process, and we were able to run as many as eight processes on a single machine and API key without being slowed by Google’s rate limits on API requests. This gave a total effective road-speed of 500–600 km/h. Based on spot-checks against the Street View browser interface, we estimate that we discovered over 99% of the available current panoramas in this way. For the 63 districts of the five provinces examined in this paper, this amounted to just over 2.5 million panoramas.
To restrict to homegardens, we downloaded images from only those panoramas whose locations were within 10 m of the center of a “settled” cell of the High Resolution Settlement Layer (HRSL) (Facebook Connectivity Lab and Center for International Earth Science Information Network Columbia University 2016): steps 2 and 3 in Fig. 2. This is a worldwide grid of cells, approximately 30 m across, that have been classified as “settled” or “not settled” based on automated analysis of satellite imagery. For each district we either randomly selected 3000 such panoramas, or chose all of them if there were fewer than 3000. The resulting stratified random sample of approximately 150,000 panoramas contains views of approximately one tenth of all homesteads in the five-province region, based on a total population of 3.8 million and the national average household size of 3.1 persons (National Statistical Office 2011).
We determined the compass heading of the road(s) at each selected panorama by clustering the directions of nearby panoramas, and used a script-driven headless browser to download a 3840 × 2160 pixel view from the panorama facing perpendicularly to the road (randomly selecting which side), with a 90-degree horizontal field of view. We also similarly chose and downloaded an additional 4000 images to be used for training and testing of the network. We ensured that the 4000 training/testing images encompassed multiple seasons of image acquisition so that we captured seasonal variations in appearance (such as flowering vs. non-flowering mango trees) and lighting conditions (sunny and overcast, in particular).
Image tagging
We chose 11 species of plants that are commonly grown in homegardens in Thailand and that were not difficult for our team of non-botanists to recognize. These were: 1) banana (Musa sapientum), 2) coconut (Cocos nucifera), 3) Dracaena fragrans, 4) galangal (Alpinia siamense), 5) jackfruit (Artocarpus heterophyllus), 6) mango (Mangifera indica), 7) papaya (Carica papaya), 8) star gooseberry (Phyllanthus acidus), and 9–11) a group of three species of shrubs with bipinnate leaves comprising cha-om (Acacia pennata), river tamarind (Leucaena leucocephala) and Pride of Barbados (Caesalpinia pulcherrima), which were lumped into a single class because it was initially difficult for us to tell them apart. Mango, because of its significant phenological changes (prolific blossoms), was actually split into two classes (mango in bloom, and not) for training and detection. This set of 11 species accounts for almost half of the most common species found in a small-scale but highly detailed inventory of homegarden species conducted at several sites in Phrae province by Lattirasuvan et al. (2010).
We attempted to visually detect and manually outline every instance of each of these classes in the set of training and test images (step 4 in Fig. 2). In Fig. 3, we show the outlines (white curves) drawn on one example image from the test set. We found it to be cognitively least taxing to tag just a single species on each pass through the image set. For quality control, each task was performed by two different taggers, and discrepancies were resolved by the first author. Figure 3 also shows the white rectangular bounding boxes of the human-drawn outlines. These boxes are what was used to train the network.
In an image from the test set, we can compare the network-detected objects (colored boxes) and the human-identified objects (white outlines and bounding boxes). A mango tree, two coconut palms, a dracaena plant, and a jackfruit tree are correctly detected and well localized by the network. A spurious coconut palm detection occurs on the right. Latitude 18.320885°N, longitude 100.316757°E. Base image copyright Google, Inc., 2019. Link: https://www.google.com/maps/@18.3208851,100.3167573,3a,90y,129.11h,93.19t/data=!3m6!1e1!3m4!1sLE0_Z9GS6V_ojV23Oxi6eA!2e0!7i13312!8i6656
Network training and object detection
We initially experimented with two different convolutional neural networks (CNNs), RetinaNet and YOLOv3 (Lin et al. 2018), for object detection and chose RetinaNet because it greatly outperformed YOLOv3 in our trials. We used the implementation available at https://github.com/fizyr/keras-retinanet, which we modified to accommodate the input and output formats requisite for our other scripts. We used the ResNet-50 network as the backbone, training for 50 epochs with 10,000 steps per epoch, and using an initial learning rate of 10−5 (step 5 in Fig. 2). This took two days using an Nvidia Titan V GPU. The objects detected by the trained CNN in the example image of Fig. 3 are drawn in color and annotated with the network’s confidence level in the detection, which can range from 0 (“no confidence”) to 1 (“certainty”).
Of the 4000 tagged images, 350 were set aside for testing and not used during the training of the CNN. After training, we applied the network to the images in the test set (step 6 in Fig. 2). To measure its effectiveness, we calculated the average precision, APIoU = 0.5, on the test set for each species. This metric, and its many variants, have become widely accepted as benchmarks of detection network performance since its inclusion in the PASCAL Visual Object Classes (VOC) Challenge (Everingham et al. 2010). Average precision (AP) summarizes the relationship between precision = # true detections/(# true detections + # false detections) and recall = # true detections/(# true detections + # undetected instances). Specifically it is the area under the least non-increasing upper bound of the precision-recall curve, which is parametrized by confidence level. Its maximum possible, and ideal, value is 1. In computing the AP, a detection is characterized as true if the predicted box and the ground-truth box overlap to the extent that the area of their intersection divided by that of their union (Intersection over Union, IoU) is no less than some chosen threshold. We chose 0.5 for this threshold. While IoU = 0.5 may be considered low in some applications (such as a self-driving car trying to avoid a pedestrian), it is quite acceptable in ours, where merely detecting the presence of an instance is the primary concern, rather than localizing it very precisely in the image. The mean average precision (mAP) is the AP averaged over all species.
Application of the CNN and visualization of the results
The trained CNN was finally applied to the remaining set of approximately 150,000 images (step 7 in Fig. 2). To visualize the output of the network, we used kernel density estimation (KDE) to compute local averages of quantities such as the prevalence of each species and the number of species seen in each image (step 8 in Fig. 2). For each point p in a dense lattice of locations in the region, we evaluated the formula
where fi is the value of the quantity of interest in the i th image, pi is the location of the i th image, and K is the KDE kernel, for which we chose a two-dimensional Gaussian function of the distance between the evaluation point p and the location pi of the i th image. Thus KDEf(p) is an average of the fi weighted by their proximity to point p.
The standard deviation of the Gaussian kernel, or bandwidth, was chosen based on the distance scale we wanted to average over. For a map of the prevalence of a particular species, fi was taken to be 1 if the species were present in image i, and 0 if not. For a map depicting the average number of target species detected in each image, fi is the number of distinct species detected in image i. These local averages were visualized by creating a map overlay in which the color corresponds to the value of KDEf. Although a value of KDEf is defined at every point in the study region, it is of little significance in places far from any image locations: therefore the opacity of the overlay was set to a Gaussian function of the distance to the closest image location, with the same standard deviation as the KDE kernel, so that the overlay is visible only where it is relevant.
Results
CNN performance
The AP of the CNN on the test set for most of our target species is satisfactory, as shown in Table 1. Our mAP is at the top of the range of mean average precision of between 0.48 and 0.55 obtained with the same network on the COCO benchmark dataset (Lin et al. 2018). Moreover, jackfruit is the only species for which the AP is significantly below this range, which can likely be attributed to its low prevalence in the training set. The number of instances of each species which were found in the training set is shown in Table 1. We omitted the bipinnate-leaved shrub group (cha-om, river tamarind, and Pride of Barbados) and galangal at this point in our analysis because the false positive rates were deemed too high at reasonable values of detection rates. This brought our number of classes to seven.
To provide a more detailed picture of the performance of the network that is pertinent to the particular use of the network output we are making in the current paper, in Fig. 4 we treat the network as a classifier of images - as positive or negative for the presence of each species - and display the numbers of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) as a function of confidence threshold. These plots illustrate the compromise that must be found between high specificity = TN/(TN + FP), which is achieved with high confidence thresholds, and high sensitivity TP/(TP + FN), which is achieved with low confidence thresholds. We hoped to find a choice of confidence threshold at which true positives and true negatives (blues in the figure) predominate. Acceptable compromises are seen to exist for all species except perhaps jackfruit, presumably due to the small number of examples in the training data as noted above. For simplicity, the confidence threshold was set at 0.5 for all species when generating the heatmaps that follow.
Treating the network as a classifier of the images as positive or negative for the presence of each species, these plots show the numbers of images (vertical axis) in the test set that are true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) for each of the seven species, as a function of network confidence threshold
Application of trained CNN
In this subsection, we present heatmaps that illustrate patterns of homegardening practices measured by our survey spanning a large portion of northern Thailand. Our technique of fine-grained observations carried out over a large geographic region reveals patterns that might not emerge from studies of small numbers of gardens. The heatmap in Fig. 5 shows with color the local average of the number of target species detected in each image, a metric that for brevity we will refer to henceforth as the intra-garden variety, over almost the whole 5-province study area. For this regional picture, we perform local spatial averaging with a KDE bandwidth of 2.5 km. It is seen that the large city of Chiang Mai and its surroundings constitutes a region tens of kilometers across where the intra-garden variety is low (less than around one species in each image) compared with the more rural provinces to the east, where the intra-garden variety typically ranges between 1.5 and 2 species in each image.
Heatmap of “intra-garden variety” (locally averaged number of target species detected in each image) with KDE bandwidth 2.5 km. Almost the entire 5-province study-area is shown. Black dots mark the sampled panorama locations: approximately 3000 per district (amphoe), totaling about 150,000. The opacity of the overlay is a Gaussian function of distance to the nearest panorama with standard deviation also 2.5 km. The two white rectangles at the upper right mark the regions shown in Figs. 6 and 7. The inset in the lower right corner is a histogram of cell intra-garden variety. Background map tiles copyright Stamen Design (CCby3.0)
A similar effect, though less extreme and much less extensive, is seen at the other four provincial capitals, which are indicated by arrows in the figure. Another prominent feature of this heatmap is the relatively low intra-garden variety over almost the entire western edge of our study area: this is addressed in the Discussion section.
Each of these features suggests phenomena that could be examined further using either this method or traditional approaches to homegarden study. The histogram inset at the lower right of Fig. 5 shows the frequency distribution of the intra-garden variety in this map. Coarsely, it is unimodal with some excesses near rational numbers that arise from cells where only a small number of images contribute significantly to the local weighted average. Only a small fraction of cells have a local average higher than two detected species in each image with this 2.5 km-bandwidth smoothing, but higher averages occur at finer spatial scales as seen in Fig. 6.
A heatmap of intra-garden variety (locally averaged number of target species detected in each image) at higher resolution than in Fig. 5, using a KDE bandwidth of 500 m. High village-to-village variation is evident. The region shown here is indicated by the smaller box drawn at the upper right of Fig. 5. Each black dot marks a sampled panorama location. The color scale is the same as in Fig. 5. Background map tiles copyright Stamen Design (CCby3.0)
Figure 6 is a higher-resolution heatmap - with 500 m KDE bandwidth - of the intra-garden variety, covering a region indicated by the smaller of the two boxes at the upper right of Fig. 5. It reveals finer kilometer-scale structure that is largely smoothed out in the multi-province picture of Fig. 5. In Fig. 6, we see that neighboring villages have quite different intra-garden variety, with examples at 1.25, 1.75 and almost 2.5 species in each image on average. Also, visually prominent is a development of some kind in the center of the picture where, along two long straight parallel roads, intra-garden variety is very low: at or below about 0.5 species in each image on average.
In Fig. 7, we show the prevalence of individual species, at the same 500 m kernel bandwidth used in Fig. 6, over the region marked by the larger of the two rectangles at the upper right of Fig. 5. In each panel of Fig. 7, the color denotes the local average fraction of images in which the particular species was detected. It is immediately apparent that coconut and banana were the most frequently detected species. Both show substantial heterogeneity, with low frequency in the core of the town of Chiang Kham (close to the center of each images in Fig. 7). Coconut attains its highest prevalence immediately outside the town, while banana appears to be most prevalent in the parts most remote from the town. The prevalence of dracaena persists right into the core of Chiang Kham while that of star gooseberry does not. Likewise mango, which is the most homogeneously distributed of these six species, remains common in the core of the town, while papaya is markedly rarer there than in the surrounding villages.
For six species, heatmaps of their local prevalence - the local average fraction of images in which the species is detected - in the vicinity (larger white rectangle at upper right in Fig. 5) of the town of Chiang Kham (white arrow in banana panel), Phayao province. A Gaussian KDE with bandwidth 500 m was used. Although all degrees of prevalence between 50% and 100% are colored yellow, few cells have a banana prevalence above 50%, and few have a coconut prevalence above 60%. Background map tiles copyright Stamen Design (CCby3.0)
Discussion
As we have just described, our survey reveals significant variations in horticultural activity at both examined spatial scales. The blue area on the left of Fig. 5, indicating lower numbers of species in each image along the western edge of the study area, is among the most striking features of that figure. Since this area is roughly the edge of the portion of Thailand settled by Karen people originally from Burma (Delang 2003), our guess is that we are detecting ethnic differences in horticultural practice (Srithi et al. 2012). But the literature offers many other variables that can drive or influence homegarden plant selection and intra-garden variety: garden owner income and other socioeconomic factors (Gajaseni and Gajaseni 1999; Galluzzi et al. 2010; Pandey et al. 2007; Wezel and Bender 2003), proximity to a central city (Huai et al. 2011), climate characteristics (Huai et al. 2011), the utility of a specific crop (Gajaseni and Gajaseni 1999), topography, soil properties, and so on. A comparison of medicinal plant usage among Thai ethnic groups found that there was more similarity in medicinal plant species usage among villages within same region than there was similarity in medicinal plant species usage within a given ethnic group (Phumthum and Balslev 2019). Further, Phumthum and Balslev call for ethnobotanical research that is regional because studies of individual villages may, it finds, miss these larger trends. Indeed, the high village-to-village variation observed here backs this up, illustrating a potential hazard of drawing broad conclusions from solely the kind of small-scale sampling used in traditional studies. The data we generate can clearly support studies of regional scale. We do not attempt here to determine the meaning of the patterns of homegardening we have detected - we are not well qualified to do so - but we believe our approach offers a new way of obtaining information for the investigation of those connections that is complementary to traditional approaches employing small samples examined intensively. Our technique also has some limitations and challenges, which we now enumerate.
The intra-garden variety here is strictly with respect to the species we have trained our CNN to recognize: if gardeners in a district prefer a species that is not on our list over one on the list (Dimocarpus longan over mango, for example), all other things being equal, that would register here as a lower local average number of species in each image. Such a preference could in fact be responsible for the “blue” western edge of Fig. 5. At any stage of its development, our tool will only have the ability to identify plants in a prescribed set of species. Surveys conducted with it are thus quite different from traditional ones described in the literature (such as Lattirasuvan et al. (2010)) that attempt to generate an exhaustive inventory of species grown in a small sample of gardens: ours is in a sense the opposite - spatially exhaustive for a restricted set of species.
There can be intrinsic difficulties in distinguishing and identifying plant species visually from macroscopic images, even of high resolution. While some species, such as papaya, are highly distinctive and hard to confuse with anything else, others belong to groups of visually similar species (lemongrass among other grasses, for example) that are less easily distinguishable both to the human eye and to a CNN. It is for this reason that we have not included results on galangal or the group of shrubs containing cha-om in this paper; with the amount of training data we have prepared so far, our false-positive rates were too high. For these species in particular, we judge that good results can be achieved if the quantity of training data can be doubled or quadrupled. But for other species, achieving reliable automated detection from GSV-like images may be impossible or require a prohibitive amount of labor to compile adequate training data. Vegetables, in particular, small and low to the ground, may pose a great challenge.
We note that while our measured variations in individual species prevalence are meaningful, the absolute prevalences of different species should be compared only with caution and after careful calibration, because the detectability radius can vary considerably among species. Coconut palms, in particular, because they are tall, are detected even when behind buildings, walls, and trees that would obscure instances of most other species on our list. There is a potential for some double-counting of instances of species like coconut by seeing them both at a distance and close up in different images. This can be limited by rejecting detected instances whose bounding box is smaller than a suitably chosen threshold size.
There are also limitations that result from using Google Street View imagery in particular. Firstly, we do not have any control over its collection. Spatially, we have imagery only from where the GSV cars have traveled, though fortunately in Thailand this does include a very large fraction of all roads, even down to unpaved single-lane tracks. Secondly, even where we do have imagery, we can see only plants with an unobstructed line of sight from the street. The GSV camera is quite high off the ground - about 2.5 m - allowing us to see over most fences and garden walls.. Temporally, most locations have imagery only from a single date, so the opportunities for exact simultaneous comparisons and for longitudinal studies are limited.
Finally, in the present work, we have relied on the HRSL classification of locations as settled or not settled to restrict our attention to images of homesteads. The HRSL classification is not perfect. We have observed, for example, that an HRSL cell containing an isolated (uninhabited) shelter in an agricultural crop field is typically falsely labeled as settled. To prevent such misidentifications from distorting the heatmaps presented here, we omitted isolated panoramas that did not have at least two other panoramas within a radius of two KDE bandwidths. It is possible that a second CNN, trained to recognize homesteads, could be used to filter out non-homestead imagery more accurately.
Conclusions
We believe our results show that GSV imagery combined with object detection by a CNN provides an unprecedented opportunity for large-scale, densely sampled, high-resolution surveys of some aspects of homegardening practices. We have demonstrated that the tool we have developed can already reliably detect six species of plants common in homegardens in Thailand (mango, papaya, banana, coconut, gooseberry, dracaena). It is also near the point of doing well on three other species (jackfruit, cha-om, galangal), and has the potential to be extended to others.
While the per-species labor required to tag the images used to train the detection network is not trivial, once that work is done, enormous plant surveys can then be constructed with minimal additional human labor. Even for purposes where the automated discrimination is too coarse, this approach could serve as a useful initial step in a survey with human horticultural experts then refining the labeling, such as by plant variety or cultivar. There are some constraints under which our tool must operate, but we anticipate that it can be deployed by researchers and policymakers to usefully augment available data about homegardening across large regions.
Data availability
Data is publicly available via Google Maps.
References
Berland A, Lange DA (2017) Google Street View shows promise for virtual street tree surveys. Urban For Urban Green 21:11–15. https://doi.org/10.1016/j.ufug.2016.11.006
Berti PR, Krasevec J, FitzGerald S (2004) A review of the effectiveness of agriculture interventions in improving nutrition outcomes. Public Health Nutr 7:599–609. https://doi.org/10.1079/PHN2003595
Cedar Lake Ventures (2020) https://weatherspark.com. Accessed 15 Nov 2020
Clarke LW, GDJ LL, Yu Z (2014) Drivers of plant biodiversity and ecosystem service production in home gardens across the Beijing Municipality of China. Urban Ecosyst 17:741–760. https://doi.org/10.1007/s11252-014-0351-6
Colding J, Barthel S (2013) The potential of "urban green commons" in the resilience building of cities. Ecol Econ 86:156–166. https://doi.org/10.1016/j.ecolecon.2012.10.016
Coomes OT, Ban N (2004) Cultivated plant species diversity in home gardens of an Amazonian peasant village in northeastern Peru. Econ Bot 58:420–434. https://doi.org/10.1663/0013-0001(2004)058[0420:CPSDIH]2.0.CO;2
Cruz Garcia GS, Struik PC (2015) Spatial and seasonal diversity of wild food plants in home gardens of Northeast Thailand. Econ Bot 69:99–113. https://doi.org/10.1007/s12231-015-9309-8
Damm H (2008) Topography of northern Thailand. Wikimedia Commons.https://commons.wikimedia.org/wiki/File:Lanna-Map.svg. Accessed 12 Aug 2020
Das T, Das AK (2005) Inventorying plant biodiversity in homegardens: a case study in Barak Valley, Assam, North East India. Curr Sci 89:155–163
Delang C (2003) Living at the edge of Thai society : the Karen in the highlands of northern Thailand. Routledge, London
Deus E, Silva JS, Catry FX, Rocha M, Moreira F (2016) Google Street View as an alternative method to car surveys in large-scale vegetation assessments. Environ Monit Assess 188:560. https://doi.org/10.1007/s10661-016-5555-1
Everingham M, Gool LV, Williams CKI, Winn J, Zisserman A (2010) The PASCAL Visual Object Classes (VOC) Challenge. Int J Comput Vis 88:303–338. https://doi.org/10.1007/s11263-009-0275-4
Facebook Connectivity Lab and Center for International Earth Science Information Network Columbia University (2016) High Resolution Settlement Layer (HRSL). https://ciesin.columbia.edu/data/hrsl/#data. Accessed 17 Jul 2017
Fernandes ECM, Nair PKR (1986) An evaluation of the structure and function of tropical homegardens. Agric Syst 21(4):279–310
Gajaseni J, Gajaseni N (1999) Ecological rationalities of the traditional homegarden system in the Chao Phraya Basin, Thailand. Agrofor Syst 46:3–23
Galluzzi G, Eyzaguirre P, Negri V (2010) Home gardens: Neglected hotspots of agro-biodiversity and cultural diversity. Biodivers Conserv 19:3635–3654. https://doi.org/10.1007/s10531-010-9919-5
George MV, Christopher G (2019) Structure, diversity and utilization of plant species in tribal homegardens of Kerala, India. Agrofor Syst 94:297–307. https://doi.org/10.1007/s10457-019-00393-5
Huai H, Xu W, Wen G, Bai W (2011) Comparison of the homegardens of eight cultural groups in Jinping County, Southwest China. Econ Bot 65:345–355. https://doi.org/10.1007/s12231-011-9172-1
Idohou R, Fandohan B, Salako VK, Kassa B, Gbèdomon RC, Yédomonhan H, Glèlè Kakaï RL, Assogbadjo AE (2014) Biodiversity conservation in home gardens: traditional knowledge, use patterns and implications for management. Int J Biodivers Sci Ecosyst Serv Manag 10:89–100. https://doi.org/10.1080/21513732.2014.910554
Jemal O, Callo-Concha D, van Noordwijk M (2018) Local agroforestry practices for food and nutrition security of smallholder farm households in southwestern Ethiopia. Sustainability 10:2722. https://doi.org/10.3390/su10082722
Kabir ME, Webb EL (2008) Floristics and structure of southwestern Bangladesh homegardens. Int J Biodivers Sci Manag 4:54–64. https://doi.org/10.1080/17451590809618183
Kumar BM, Nair PKR (2004) The enigma of tropical homegardens. Agrofor Syst 61-2:135–152
Lattirasuvan T, Tanaka S, Nakamoto K, Hattori D, Sakurai K (2010) Ecological characteristics of home gardens in northern Thailand. Tropics 18:171–184. https://doi.org/10.3759/tropics.18.171
Lin T-Y, Goyal P, Girshick R, He K, Dollar P, Facebook AI Research (2018) Focal Loss for Dense Object Detection
Mathewos M, Hundera K, Biber-Freudenberger L (2018) Planting Fruits and Vegetables in Homegarden as a Way to Improve Livelihoods and Conserve Plant Biodiversity. Agriculture 8. https://doi.org/10.3390/agriculture8120190
Molla A, Kewessa G (2015) Woody species diversity in traditional agroforestry practices of Dellomenna District, Southeastern Ethiopia: Implication for Maintaining Native Woody Species. Int J biodiver 2015:1–13. https://doi.org/10.1155/2015/643031
National Statistical Office of Thailand (2011) Population and Housing Census. http://web.nso.go.th. Accessed Sept 2019
Pala NA, Sarkar BC, Shukla G, Chettri N, Deb S, Bhat JA, Chakravarty S (2019) Floristic composition and utilization of ethnomedicinal plant species in home gardens of the Eastern Himalaya. J Ethnobiol Ethnomed 15:14–16. https://doi.org/10.1186/s13002-019-0293-4
Pandey CB, Rai RB, Singh L, Singh AK (2007) Homegardens of Andaman and Nicobar, India. Agric Syst 92:1–22. https://doi.org/10.1016/j.agsy.2006.01.009
Panyadee P, Balslev H, Wangpakapattanawong P, Inta A (2016) Woody Plant Diversity in Urban Homegardens in Northern Thailand. Econ Bot 70:285–302. https://doi.org/10.1007/s12231-016-9348-9
Panyadee P, Balslev H, Wangpakapattanawong P, Inta A (2018) Karen Homegardens: characteristics, Functions, and Species Diversity. Econ Bot 72:1–19. https://doi.org/10.1007/s12231-018-9404-8
Panyadee P, Balslev H, Wangpakapattanawong P, Inta A (2019) Medicinal plants in homegardens of four ethnic groups in Thailand. J Ethnopharmacol 239:111927. https://doi.org/10.1016/j.jep.2019.111927
Phumthum M, Balslev H (2019) Use of medicinal plants among thai ethnic groups: a comparison. Econ Bot 73:64–75. https://doi.org/10.1007/s12231-018-9428-0
Rayol BP, Do Vale I, Miranda IS (2019) Tree and palm diversity in homegardens in the Central Amazon. Agrofor Syst 93:515–529. https://doi.org/10.1007/s10457-017-0144-z
Ringland J, Bohm M, Baek S-R (2019) Characterization of food cultivation along roadside transects with Google Street View imagery and deep learning. Comput Electron Agric 158:36–50. https://doi.org/10.1016/j.compag.2019.01.014
Schreinemachers P et al (2015) The effect of women's home gardens on vegetable production and consumption in Bangladesh. Food Sec 7:97–107. https://doi.org/10.1007/s12571-014-0408-7
Serrano-Ysunza AA, van der Wal H, Gallardo-Cruz JA, Ramos-Munoz DE, Vaca RA (2018) A 6-year longitudinal study on agrobiodiversity change in homegardens in Tabasco, Mexico. Agrofor Syst 92:1485–1494. https://doi.org/10.1007/s10457-017-0094-5
Shumi G et al (2018) Land use legacy effects on woody vegetation in agricultural landscapes of south-western ethiopia. Divers Distrib 24:1136–1148. https://doi.org/10.1111/ddi.12754
Srithi K, Trisonthi C, Wangpakapattanawong P, Srisanga P, Balslev H (2012) Plant Diversity in Hmong and Mien Homegardens in Northern Thailand. Econ Bot 66:192–206
Tadesse E, Abdulkedir A, Khamzina A, Son Y, Noulekoun F (2019) Contrasting species diversity and values in home gardens and traditional parkland agroforestry systems in Ethiopian sub-humid lowlands. Forests 10:266. https://doi.org/10.3390/f10030266
Trinh LN, Watson JW, Hue NN, de NN, Minh NV, Chu P, Sthapit BR, Eyzaguirre PB (2003) Agrobiodiversity conservation and development in Vietnamese home gardens. Agric Ecosyst Environ 97:317–344. https://doi.org/10.1016/S0167-8809(02)00228-1
Vibhuti, Bargali K, Bargali SS (2018) Effects of homegarden size on floristic composition and diversity along an altitudinal gradient in Central Himalaya, India. Curr Sci 114(00113891):2494–2503. https://doi.org/10.18520/cs/v114/i12/2494-2503
Webb EL, Kabir ME (2009) Home gardening for tropical biodiversity conservation. Conserv Biol 23:1641–1644. https://doi.org/10.1111/j.1523-1739.2009.01267.x
Weinberger K (2013) Home and community gardens in Southeast Asia: potential and opportunities for contributing to nutrition-sensitive food systems. Food Sec 5:847–856. https://doi.org/10.1007/s12571-013-0299-z
Wezel A, Bender S (2003) Plant species diversity of homegardens of Cuba and its significance for household food supply. Agrofor Syst 57:39–49
Whitney CW, Luedeling E, Hensel O, Tabuti JRS, Krawinkel M, Gebauer J, Kehlenbeck K (2018) The role of homegardens for food and nutrition security in Uganda. Hum Ecol 46:497–514. https://doi.org/10.1007/s10745-018-0008-9
Woldeamanual W, Asfaw Z, Lemessa D (2018) The effect of the surrounding landscape and socioeconomic characteristics on woody species diversity in homegardens of Shewarobit District, Northeast Ethiopia. Small-Scale Forestry 17:471–483. https://doi.org/10.1007/s11842-018-9398-1
Yamane Y, Kularatne J, Ito K (2018) Diversity of cropping patterns and factors affecting homegarden cultivation in Kiboguwa on the eastern slopes of the Uluguru mountains in Tanzania. Agriculture 8:141. https://doi.org/10.3390/agriculture8090141
Zuanzuanfuwa (2012) Orthographic map of Thailand. Wikimedia Commons.https://commons.wikimedia.org/wiki/File:Thailand_(orthographic_projection).svg. Accessed 12 Aug 2020
Acknowledgements
The authors gratefully acknowledge the assistance of our student research assistants: Matthew Straub, Nicholas Lahue, Kevin Medina, and Niranjan Ravichandra. We also benefited from the expertise of Nirin Prasirthang, our Thai horticultural consultant. This project was supported by a grant from the Community for Global Health Equity at the University at Buffalo.
Funding
This project was supported by a grant from the Community for Global Health Equity at the University at Buffalo.
Author information
Authors and Affiliations
Contributions
Conception and design: JR, MB, SB.
Acquisition and curation of data: JR.
Image tagging: JR, MB, SB, ME, and student assistants.
Computer programing and image processing: JR, ME.
Results interpretation and manuscript writing: JR, MB, SB, ME.
Corresponding author
Ethics declarations
Conflict of interest
Not applicable.
Code availability
Upon request from corresponding author.
Additional information
Communicated by: H. Babaie
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ringland, J., Bohm, M., Baek, SR. et al. Automated survey of selected common plant species in Thai homegardens using Google Street View imagery and a deep neural network. Earth Sci Inform 14, 179–191 (2021). https://doi.org/10.1007/s12145-020-00557-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12145-020-00557-3