Abstract
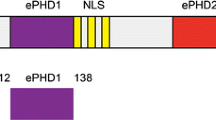
Phafin2 is a peripheral protein that triggers cellular signaling from endosomal and lysosomal compartments. The specific subcellular localization of Phafin2 is mediated by the presence of a tandem of phosphatidylinositol 3-phosphate (PtdIns3P)-binding domains, the pleckstrin homology (PH) and the Fab-1, YOTB, Vac1, and EEA1 (FYVE) domains. The requirement for both domains for binding to PtdIns3P still remains unclear. To understand the molecular interactions of the Phafin2 PH domain in detail, we report its nearly complete 1H, 15N, and 13C backbone resonance assignments.

Similar content being viewed by others
Data availability
The assignments of resonances of the Phafin2 PH domain were deposited in the Biological Magnetic Resonance Bank database under accession number 26309.
References
Chen W, Li N, Chen T, Han Y, Li C, Wang Y, He W, Zhang L, Wan T, Cao X (2005) The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal-mitochondrial pathway. J Biol Chem 280:40985–40995
Choi AM, Ryter SW, Levine B (2013) Autophagy in human health and disease. N Engl J Med 368:651–662
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Feng JR, He L, Li YQ, Xiao F, Hu G (2019) Modeling of PH domains and phosphoinositides interactions and beyond. Protein Rev—Purinergic Recept 20(1111):19–32
Gailite I, Egger-Adam D, Wodarz A (2012) The phosphoinositide-associated protein rush hour regulates endosomal trafficking in Drosophila. Mol Biol Cell 23:433–447
Grzesiek S, Anglister J, Bax A (1993) Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson 101:114–119
Hirata N, Suizu F, Matsuda-Lennikov M, Tanaka T, Edamura T, Ishigaki S, Donia T, Lithanatudom P, Obuse C, Iwanaga T et al (2018) Functional characterization of lysosomal interaction of Akt with VRK2. Oncogene 37:5367–5386
Hyberts SG, Takeuchi K, Wagner G (2010) Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc 132:2145–2147
Jiang Z, Liang Z, Shen B, Hu G (2015) Computational analysis of the binding specificities of PH domains. Biomed Res Int 2015:792904
Kariuki SN, Ghodke-Puranik Y, Dorschner JM, Chrabot BS, Kelly JA, Tsao BP, Kimberly RP, Alarcon-Riquelme ME, Jacob CO, Criswell LA et al (2015) Genetic analysis of the pathogenic molecular sub-phenotype interferon-alpha identifies multiple novel loci involved in systemic lupus erythematosus. Genes Immun 16:15–23
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327
Lemmon MA, Ferguson KM, Schlessinger J (1996) PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell 85:621–624
Lin WJ, Yang CY, Lin YC, Tsai MC, Yang CW, Tung CY, Ho PY, Kao FJ, Lin CH (2010) Phafin2 modulates the structure and function of endosomes by a Rab5-dependent mechanism. Biochem Biophys Res Commun 391:1043–1048
Maciejewski MW, Schuyler AD, Gryk MR, Moraru II, Romero PR, Ulrich EL, Eghbalnia HR, Livny M, Delaglio F, Hoch JC (2017) NMRbox: a resource for biomolecular NMR computation. Biophys J 112:1529–1534
Manning BD, Toker A (2017) AKT/PKB signaling: navigating the network. Cell 169:381–405
Matsuda-Lennikov M, Suizu F, Hirata N, Hashimoto M, Kimura K, Nagamine T, Fujioka Y, Ohba Y, Iwanaga T, Noguchi M (2014) Lysosomal interaction of Akt with Phafin2: a critical step in the induction of autophagy. PLoS ONE 9:e79795
Montgomerie S, Cruz JA, Shrivastava S, Arndt D, Berjanskii M, Wishart DS (2008) PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res 36:W202–W209
Muhandiram DR, Kay LE (1994) Gradient-enhanced triple-resonance 3-dimensional Nmr experiments with improved sensitivity. J Magn Reson 103:203–216
Pedersen NM, Raiborg C, Brech A, Skarpen E, Roxrud I, Platta HW, Liestol K, Stenmark H (2012) The PtdIns3P-binding protein Phafin 2 mediates epidermal growth factor receptor degradation by promoting endosome fusion. Traffic 13:1547–1563
Shamsara E, Shamsara J (2020) Bioinformatics analysis of the genes involved in the extension of prostate cancer to adjacent lymph nodes by supervised and unsupervised machine learning methods: the role of SPAG1 and PLEKHF2. Genomics 112:3871–3882
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Tang TX, Jo A, Deng J, Ellena JF, Lazar IM, Davis RM, Capelluto DG (2017a) Structural, thermodynamic, and phosphatidylinositol 3-phosphate binding properties of Phafin2. Protein Sci 26:814–823
Tang TX, Xiong W, Finkielstein CV, Capelluto DGS (2017b) Identification of lipid binding modulators using the protein-lipid overlay assay. Methods Mol Biol 1647:197–206
Tang TX, Finkielstein CV, Capelluto DGS (2020) The C-terminal acidic motif of Phafin2 inhibits PH domain binding to phosphatidylinositol 3-phosphate. Biochim Biophys Acta Biomembr 1862:183230
Ying J, Delaglio F, Torchia DA, Bax A (2017) Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J Biomol NMR 68:101–118
Acknowledgements
We thank Dr. Janet Webster for assistance during preparation of the manuscript. This work was funded by the 4-VA Collaborative Research Program and by the Virginia Academy of Science Mary Louis Andrews for Cancer Research (to D.G.S.C.). T.T. was supported by the Joseph Frank Hunkler Memorial Fellowship. Some of the NMR data presented in this work was collected using an NMR instrument purchased with an NIH High End Instrumentation grant (S10 RR023035), which is housed in the Biomolecular Magnetic Resonance Facility at the University of Virginia.
Author information
Authors and Affiliations
Contributions
JFE and DGSC, research design; TT, purified recombinant protein; JFE, TT, and NS, performed and analyzed two-dimensional NMR experiments; JFE, performed and analyzed three-dimensional NMR experiments; DGSC, wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ellena, J.F., Tang, TX., Shanaiah, N. et al. Backbone 1H, 15N, and 13C resonance assignments of the Phafin2 pleckstrin homology domain. Biomol NMR Assign 16, 27–30 (2022). https://doi.org/10.1007/s12104-021-10054-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-021-10054-3