Abstract
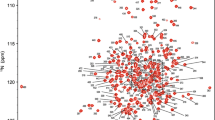
The forkhead-associated (FHA) domain is known as a phosphopeptide recognition domain embedded in regulatory proteins from both eukaryotes and bacteria with various biological functions. In this study, the gene encoding a predicted FHA domain from Mb1858 (residues V24-D155 from the 162 amino acid protein Mb1858) in Mycobacterium bovis was cloned, and U-13C/15N-labeled protein was prepared for backbone and side chain resonance assignments by NMR spectroscopy. These assignments are preliminary work towards the determination of the structure and phosphopeptide-binding properties using NMR methods, which will provide useful information about the function of Mb1858 protein.


Similar content being viewed by others

References
Almawi AW, Matthews LA, Guarne A (2016) FHA domains: phosphopeptide binding and beyond. Prog Biophys Mol Biol. doi:10.1016/j.pbiomolbio.2016.12.003
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe—a multidimensional spectral processing system based on Unix pipes. J Biomol NMR 6:277–293
Durocher D, Jackson SP (2002) The FHA domain. FEBS Lett 513:58–66
Goddard TD, Kneller DG (2008) SPARKY 3. University of California, San Fransisco
Hofmann K, Bucher P (1995) The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci 20:347–349
Hong WL, Yu XW, Li CM, Xie JP (2013) Function and evolution of ubiquitous bacterial signaling adapter phosphopeptide recognition domain FHA. Cell Signal 25:660–665
Liu BA, Engelmann BW, Nash PD (2012) The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett 586:2597–2605
Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. BioChemistry 28:7510–7516
O’Reilly LM, Daborn CJ (1995) The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber Lung Dis 76(Suppl 1):1–46
Obsilova V, Kopecka M, Kosek D, Kacirova M, Kylarova S, Rezabkova L, Obsil T (2014) Mechanisms of the 14-3-3 protein function: regulation of protein function through conformational modulation. Physiol Res 63(Suppl 1):S155-164
Pallen M, Chaudhuri R, Khan A (2002) Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol 10:556–563
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320-324
Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, Janjua H, Jiang M, Kornhaber GJ, Lee DY, Locke JY, Ma LC, Maglaqui M, Mao L, Mitra S, Patel D, Rossi P, Sahdev S, Sharma S, Shastry R, Swapna GV, Tong SN, Wang D, Wang H, Zhao L, Montelione GT, Acton TB (2010) The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol 172:21–33
Acknowledgements
We thank for grant supports from Protein Structure Initiative-Biology Program by the National Institute of General Medical Sciences (Grant Nos: U54-GM074958 and U54-GM094597), the “Hundred Talents Program” of Chinese Academy of Sciences, the Ministry of Science and Technology of China (Grant No: 2016YFA051201) and the National Natural Science Foundation of China (Grant No: 21575155).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
He, T., Li, S., Hu, R. et al. Chemical shift assignments of Mb1858 (24-155), a FHA domain-containing protein from Mycobacterium bovis . Biomol NMR Assign 12, 1–4 (2018). https://doi.org/10.1007/s12104-017-9769-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-017-9769-0