Abstract
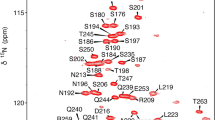
The protein SP_0782 from Streptococcus pneumonia is a small homodimeric protein that belongs to a protein family containing representative members with single-stranded DNA (ssDNA) binding functions. The ssDNA binding of the homolog YdbC from Lactococcus lactis was previously characterized when bound to a 20-mer of pyridine-rich ssDNA, sharing an overall similar structural fold with the human transcription coactivator PC4. We report that SP_0782 exhibits distinct differences in ssDNA binding properties from YdbC as revealed by NMR titration experiments. Unlike the binding of the ssDNA dT19G1 to PC4 and YdbC, SP_0782 resulted in aggregation. In addition, SP_0782 exhibits favorable binding to shorter ssDNA such as dT6. The reason is unclear, and the SP_0782 structure–function relationship remains to be elucidated. Here, we report the complete 1H, 13C, and 15N backbone and side chain NMR assignments of SP_0782, residues 7–79.


Similar content being viewed by others
References
Bahrami A, Assadi AH, Markley JL, Eghbalnia HR (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol 5(3):e1000307
Brandsen J, Werten S, van der Vliet PC, Meisterernst M, Kroon J, Gros P (1997) C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat Struct Biol 4(11):900–903
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe—a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293
Goddard TD, Kneller DG (2008) SPARKY 3. University of California, San Fransisco
Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28(19):7510–7516
Rossi P, Swapna GV, Huang YJ, Aramini JM, Anklin C, Conover K, Hamilton K, Xiao R, Acton TB, Ertekin A, Everett JK, Montelione GT (2010) A microscale protein NMR sample screening pipeline. J Biomol NMR 46(1):11–22
Rossi P, Barbieri CM, Aramini JM, Bini E, Lee HW, Janjua H, Xiao R, Acton TB, Montelione GT (2013) Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome. Nucleic Acids Res 41(4):2756–2768
Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43(5):289–318
Werten S, Moras D (2006) A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat Struct Mol Biol 13(2):181–182
Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, Janjua H, Jiang M, Kornhaber GJ, Lee DY, Locke JY, Ma LC, Maglaqui M, Mao L, Mitra S, Patel D, Rossi P, Sahdev S, Sharma S, Shastry R, Swapna GV, Tong SN, Wang D, Wang H, Zhao L, Montelione GT, Acton TB (2010) The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol 172(1):21–33
Acknowledgments
This work was supported by Protein Structure Initiative-Biology Program by the National Institute of General Medical Sciences (Grant Numbers: U54-GM074958 and U54-GM094597), the Hundred Talent Program by Chinese Academy of Sciences, the Ministry of Science and Technology (Grant number 2016YFA051201), and the National Natural Sciences Foundation of China (Grant Number: 21575155). We thank all personnel at the Rutgers protein production facility for sample preparation and technical support. All NMR data collection was conducted at the Ohio Biomedicine Center of Excellence in Strucstural Biology and Metabonomics at Miami University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, S., Ramelot, T.A., Kennedy, M.A. et al. Chemical shift assignments of the homodimer protein SP_0782 (7–79) from Streptococcus pneumoniae . Biomol NMR Assign 10, 341–344 (2016). https://doi.org/10.1007/s12104-016-9697-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-016-9697-4