Abstract
Background
Dystrophinopathies are common X-linked recessive neuromuscular disorders caused by pathogenic variants in the dystrophin gene (DMD). Analysis of the mutational spectrum in the Indian patients would be useful for confirming the diagnosis, provide genetic counseling, offer reproductive options, and importantly to determine the eligibility for the mutation-specific therapies currently approved/or undergoing trials, such as skipping of specific exons or read-through of stop codon.
Methods
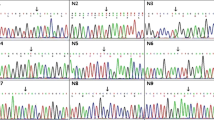
In 1660 patients diagnosed as Duchenne muscular dystrophy (DMD) /Becker muscular dystrophy (BMD) deletion- duplication analysis of all 79 exons was carried out using Multiplex ligation-dependent probe amplification (MLPA) technology. In 63 patients where no mutations were detected by MLPA, the nucleotide sequence of the DMD gene was determined by next gene sequencing. In seven cases where MLPA showed deletion of a single exon, and amplification of the specific exon was successful by polymerase chain reaction (PCR), Sanger sequencing of the concerned region was carried out to detect changes in the sequence.
Results
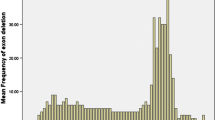
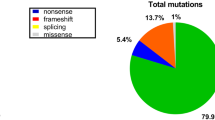
The mutation spectrum of 1660 patients with DMD/BMD was determined and 1188 (71.6%) patients were identified to have deletions or duplications of one or more exons. Of these, 1090 (65.7%) had true deletions of exons and 98 (5.9%) had duplications of exons. The most frequent change was the deletion of exon 45 (66/1090, 6.1%) and duplication of exon 2 (1/98, 11.2%). Sequencing of dystrophin gene was performed in 70 cases, and variants were identified in 68 patients (97.1% of those analyzed). Stop codon variants were observed in 34 (50%) patients, missense variants in 4 (5.9%), small deletions in 19 (27.9%), small insertions in 6 (8.8%) and slice site variants in 5 (7.4%) patients. Thirty one of 68 variants (45.5%) were novel.
Conclusions
The authors highlight the importance of identifying the type of mutation in patients with DMD. Based on the results, it is estimated that 681 (54.2%) of 1256 patients in this cohort would benefit from the currently ongoing mutation-specific therapies.






Similar content being viewed by others
References
Kingston HM, Thomas NS, Pearson PL, Sarfarazi M, Harper PS. Genetic linkage between Becker muscular dystrophy and a polymorphic DNA sequence on the short arm of the X chromosome. J Med Genet. 1983;20:255–8.
Davies KE, Pearson PL, Harper PS, et al. Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of human X chromosome. Nucleic Acids Res. 1983;11:2303–12.
Emery AE. Population frequencies of inherited neuromuscular diseases: a world survey. Neuromuscul Disord. 1991;1:19–29.
Den Dunnen JT, Grootscholten PM, Dauwerse JG, et al. Reconstruction of the 2.4 Mb human DMD gene by homologous YAC recombination. Hum Mol Genet. 1992;1:19–28.
Hoffman EP, Brown RH Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–28.
Jung D, Yang B, Meyer J, Chamberlain JS, Campbell KP. Identification and characterization of the dystrophin anchoring site on β-dystroglycan. J Biol Chem. 1995;270:27305–10.
Flanigan KM, Dunn DM, Von Niederhausern A, et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30:1657–66.
Koenig M, Beggs AH, Moyer M, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506.
Den Dunnen JT, Grootscholten PM, Bakker E, et al. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989;45:835–47.
Hu XY, Ray PN, Murphy EG, Thompson MW, Worton RG. Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin, and phenotype genotype correlation. Am J Hum Genet. 1990;46:682–95.
Roberts RG, Bobrow MA, Bentley DR. Point mutations in the dystrophin gene. Proc Natl Acad Sci U S A. 1992;89:2331–5.
Lalic T, Vossen RH, Coffa J, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13:1231–4.
Kohli S, Saxena R, Thomas E, Singh J, Verma IC. Gene changes in Duchenne muscular dystrophy: comparison of multiplex PCR and multiplex ligation-dependent probe amplification techniques. Neurol India. 2010;58:852–6.
Deepha S, Vengalil S, Preethish-Kumar V, et al. MLPA identification of dystrophin mutations and in silico evaluation of the predicted protein in dystrophinopathy cases from India. BMC Med Genet. 2017;18:67.
Vengalil S, Preethish-Kumar V, Polavarapu K, et al. Duchenne muscular dystrophy and Becker muscular dystrophy confirmed by multiplex ligation-dependent probe amplification: genotype-phenotype correlation in a large cohort. J Clin Neurol. 2017;13:91–7.
Manjunath M, Kiran P, Preethish-Kumar V, Nalini A, Singh RJ, Gayathri N. A comparative study of mPCR, MLPA, and muscle biopsy results in a cohort of children with Duchenne muscular dystrophy: a first study. Neurol India. 2015;63:58–62.
Verma PK, Dalal A, Mittal B, Phadke SR. Utility of MLPA in mutation analysis and carrier detection for Duchenne muscular dystrophy. Indian J Hum Genet. 2012;18:91–4.
Dastur RS, Kachwala MY, Khadilkar SV, Hegde MR, Gaitonde PS. Identification of deletions and duplications in the Duchenne muscular dystrophy gene and female carrier status in western India using combined methods of multiplex polymerase chain reaction and multiplex ligation-dependent probe amplification. Neurol India. 2011;59:803–9.
Sakthivel Murugan SM, Arthi C, Thilothammal N, Lakshmi BR. Carrier detection in Duchenne muscular dystrophy using molecular methods. Indian J Med Res. 2013;137:1102–10.
Swaminathan B, Shubha GN, Shubha D, et al. Duchenne muscular dystrophy: a clinical, histopathological and genetic study at neurology tertiary care center in southern India. Neurol India. 2009;57:734–8.
Aartman-Rus A, Fokkema I, Verschuuren J, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–9.
Van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–86.
Wilton SD, Fletcher S. Exon skipping and Duchenne muscular dystrophy: hope, hype and how feasible? Neurol India. 2008;56:254–62.
Roest PAM, Roberts RG, Van Der Tuijn AC, Heikoop JC, Van Ommen GJB, Den Dunnen JT. Protein truncation test (PTT) to rapidly screen the DMD-gene for translation-terminating mutations. Neuromuscul Disord. 1993;3:391–4.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215.
Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–56.
Beggs AH, Koenig M, Boyce FM, Kunkel LM. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86:45–8.
Den Dunnen JT. Leiden Muscular Dystrophy pages. Center for Human and Clinical Genetics, Leiden University Medical Center. DNA-based diagnostic techniques for DMD/BMD Deletion Detection using multiplex PCR [last updated on 2005 Mar 16]. Available at: http://www.dmd.nl/. Accessed 4 Sept 2019.
Schouten JP, McElgunn CJ, Waayer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57.
www.thermofischerscientific.com. Accessed 4 Sept 2019.
Lek M, Karczewski K, Minikel E, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Béroud C, Hamroun D, Collod-Béroud G, Boileau C, Soussi T, Claustres M. UMD (universal mutation database): 2005 update. Hum Mutat. 2005;26:184–91.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Tallapaka K, Ranganath P, Ramachandran A, et al. Molecular and histopathological characterization of patients presenting with the Duchenne muscular dystrophy phenotype in a tertiary care center in southern India. Indian Pediatr. 2019;56:556–9.
Singh B, Mandal K, Lallar M, et al. Next generation sequencing in diagnosis of MLPA negative cases presenting as Duchenne/Becker muscular dystrophies. Indian J Pediatr. 2018;85:309–10.
Ma P, Zhang S, Zhang H, et al. Comprehensive genetic characteristics of dystrophinopathies in China. Orphanet J Rare Dis. 2018;13:109.
Okubo M, Goto K, Komaki H, et al. Comprehensive analysis for genetic diagnosis of Dystrophinopathies in Japan. Orphanet J Rare Dis. 2017;12:149.
Bello L, Morgenroth LP, Gordish-Dressman H, Hoffman EP, McDonald CM, Cirak S. DMD genotypes and loss of ambulation in the CINRG Duchenne natural history study. Neurology. 2016;87:401–9.
Bladen CL, Salgado D, Monges S, et al. The TREAT-NMD DMD global database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402.
Yang J, Li SY, Li YQ, et al. MLPA-based genotype-phenotype analysis in 1053 Chinese patients with DMD/BMD. BMC Med Genet. 2013;14:29.
Wei X, Dai Y, Yu P, et al. Targeted next-generation sequencing as a comprehensive test for patients with and female carriers of DMD/BMD: a multi-population diagnostic study. Eur J Hum Genet. 2014;22:110–8.
Acknowledgements
All the staff of Molecular Genetics, Institute of Medical Genetics and Genomics, Sir Ganga Ram Hospital, New Delhi, India.
Author information
Authors and Affiliations
Contributions
ICV: Concept and design of the work, data interpretation, and finalized the manuscript; ICV, RDP, SBM: Recruited and phenotyped the participants and performed clinical investigations; ET, KS, SK, RS: Performed molecular genetic experiments; SK: Prepared figures and wrote the draft; RS, SK: Analyzed experiments and data interpretation. All authors read and approved the final manuscript. ICV will act as Guarantor for this paper.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
The patients provided written informed consent for genetic analysis.
Conflict of Interest
None.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 153 kb)
Rights and permissions
About this article
Cite this article
Kohli, S., Saxena, R., Thomas, E. et al. Mutation Spectrum of Dystrophinopathies in India: Implications for Therapy. Indian J Pediatr 87, 495–504 (2020). https://doi.org/10.1007/s12098-020-03286-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12098-020-03286-z