Abstract
Background
Cancer-associated fibroblasts (CAFs) play a significant role in regulating the clinical outcome and radiotherapy prognosis of prostate cancer (PCa). The aim of this study is to identify CAFs-related genes (CAFsRGs) using single-cell analysis and evaluate their potential for predicting the prognosis and radiotherapy prognosis in PCa.
Methods
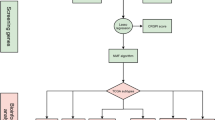
We acquire transcriptome and single-cell RNA sequencing (scRNA-seq) results of PCa and normal adjacent tissues from The GEO and TCGA databases. The "MCPcounter" and "EPIC" R packages were used to assess the infiltration level of CAFs and examine their correlation with PCa prognosis. ScRNA-seq and differential gene expression analyses were used to extract CAFsRGs. We also applied COX and LASSO analysis to further construct a risk score (CAFsRS) to assess biochemical recurrence-free survival (BRFS) and radiotherapy prognosis of PCa. The predictive efficacy of CAFsRS was evaluated by ROC curves and subgroup analysis. Finally, we integrated the CAFsRS gene signature with relevant clinical features to develop a nomogram, enhancing the predictive accuracy.
Results
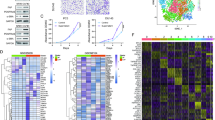
The abundance of CAFs is associated with a poor prognosis of PCa patients. ScRNA-seq and differential gene expression analysis revealed 323 CAFsRGs. After COX and LASSO analysis, we obtained seven CAFsRGs with prognostic significance (PTGS2, FKBP10, ENG, CDH11, COL5A1, COL5A2, and SRD5A2). Additionally, we established a risk score model based on the training set (n = 257). The ROC curve was used to confirm the performance of CAFsRS (The AUC values for 1, 3 and 5-year survival were determined to be 0.732, 0.773, and 0.775, respectively.). The testing set (n = 129), GSE70770 set (n = 199) and GSE116918 set (n = 248) revealed that the model exhibited exceptional predictive performance. This was also confirmed by clinical subgroup analysis. The violin plot demonstrated a statistically significant disparity in the CAFs infiltrations between the high-risk and low-risk groups of CAFsRS. Further analysis confirmed that both CAFsRS and T stage were independent prognostic factors for PCa. The nomogram was then established and its excellent predictive performance was demonstrated through calibration and ROC curves. Finally, we developed an online prognostic prediction app (https://sysu-symh-cafsnomogram.streamlit.app/) to facilitate the practical application of the nomogram.
Conclusions
The prognostic prediction risk score model we constructed could accurately predict BRFS and radiotherapy prognosis PCa, which can provide new ideas for clinicians to develop personalized PCa treatment and follow-up programs.









Similar content being viewed by others
Data availability
The scRNA-seq dataset, expression profiles and clinical information involved in this article were downloaded from TCGA Database and GEO Database. Please contact the author if you want to access the codes.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Van den Broeck T, van den Bergh R, Arfi N, Gross T, Moris L, Briers E, et al. Prognostic value of biochemical recurrence following treatment with curative intent for prostate cancer: a systematic review. Eur Urol. 2019;75(6):967–87.
Attard G, Murphy L, Clarke NW, Sachdeva A, Jones C, Hoyle A, et al. Abiraterone acetate plus prednisolone with or without enzalutamide for patients with metastatic prostate cancer starting androgen deprivation therapy: final results from two randomised phase 3 trials of the stampede platform protocol. Lancet Oncol. 2023;24(5):443–56.
Liu T, Han C, Wang S, Fang P, Ma Z, Xu L, et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J Hematol Oncol. 2019;12(1):86.
Josefsson A, Adamo H, Hammarsten P, Granfors T, Stattin P, Egevad L, et al. Prostate cancer increases hyaluronan in surrounding nonmalignant stroma, and this response is associated with tumor growth and an unfavorable outcome. Am J Pathol. 2011;179(4):1961–8.
Erdogan B, Ao M, White LM, Means AL, Brewer BM, Yang L, et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J Cell Biol. 2017;216(11):3799–816.
Nissen NI, Karsdal M, Willumsen N, et al. Collagens and cancer associated fibroblasts in the reactive stroma and its relation to cancer biology. J Exp Clin Cancer Res. 2019;38(1):115.
Zhang Y, Zhao J, Ding M, Su Y, Cui D, Jiang C, et al. Loss of exosomal mir-146a-5p from cancer-associated fibroblasts after androgen deprivation therapy contributes to prostate cancer metastasis. J Exp Clin Cancer Res. 2020;39(1):282.
Kato M, Placencio-Hickok VR, Madhav A, Haldar S, Tripathi M, Billet S, et al. Heterogeneous cancer-associated fibroblast population potentiates neuroendocrine differentiation and castrate resistance in a cd105-dependent manner. Oncogene. 2019;38(5):716–30.
Shan G, Gu J, Zhou D, Li L, Cheng W, Wang Y, et al. Cancer-associated fibroblast-secreted exosomal mir-423-5p promotes chemotherapy resistance in prostate cancer by targeting grem2 through the tgf-beta signaling pathway. Exp Mol Med. 2020;52(11):1809–22.
Hwang B, Lee JH, Bang D, et al. Single-cell rna sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1–14.
Song P, Li W, Guo L, Ying J, Gao S, He J, et al. Identification and validation of a novel signature based on nk cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma by integrated analysis of single-cell and bulk rna-sequencing. Front Immunol. 2022;13:850745.
Wang T, Dang N, Tang G, Li Z, Li X, Shi B, et al. Integrating bulk and single-cell rna sequencing reveals cellular heterogeneity and immune infiltration in hepatocellular carcinoma. Mol Oncol. 2022;16(11):2195–213.
Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218.
Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;2017(6):e26476
Aran D, Hu Z, Butte AJ, et al. Xcell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21.
Shah S, Rachmat R, Enyioma S, Ghose A, Revythis A, Boussios S, et al. Brca mutations in prostate cancer: assessment, implications and treatment considerations. Int J Mol Sci. 2021;22:23.
Kang J, La Manna F, Bonollo F, Sampson N, Alberts IL, Mingels C, et al. Tumor microenvironment mechanisms and bone metastatic disease progression of prostate cancer. Cancer Lett. 2022;530:156–69.
Wu Z, Shi J, Lai C, Li K, Li K, Li Z, et al. Clinicopathological significance and prognostic value of cancer-associated fibroblasts in prostate cancer patients. Urol Oncol. 2021;39(7):417–33.
Blom S, Erickson A, Ostman A, Rannikko A, Mirtti T, Kallioniemi O, et al. Fibroblast as a critical stromal cell type determining prognosis in prostate cancer. Prostate. 2019;79(13):1505–13.
Owen JS, Clayton A, Pearson HB, et al. Cancer-associated fibroblast heterogeneity, activation and function: implications for prostate cancer. Biomolecules. 2022;13:1.
Jimenez-Luna C, Gonzalez-Flores E, Ortiz R, Martinez-Gonzalez LJ, Antunez-Rodriguez A, Exposito-Ruiz M, et al. Circulating ptgs2, jag1, gucy2c and pgf mrna in peripheral blood and serum as potential biomarkers for patients with metastatic colon cancer. J Clin Med. 2021;10:11.
Gomez-Valenzuela F, Escobar E, Perez-Tomas R, Montecinos VP, et al. The inflammatory profile of the tumor microenvironment, orchestrated by cyclooxygenase-2, promotes epithelial-mesenchymal transition. Front Oncol. 2021;11:686792.
Zhang Z, et al. Mir-124-3p suppresses prostatic carcinoma by targeting ptgs2 through the akt/nf-kappab pathway. Mol Biotechnol. 2021;63(7):621–30.
Knuppel L, Heinzelmann K, Lindner M, Hatz R, Behr J, Eickelberg O, et al. Fk506-binding protein 10 (fkbp10) regulates lung fibroblast migration via collagen vi synthesis. Respir Res. 2018;19(1):67.
Ge Y, Xu A, Zhang M, Xiong H, Fang L, Zhang X, et al. Fk506 binding protein 10 is overexpressed and promotes renal cell carcinoma. Urol Int. 2017;98(2):169–76.
Ramadori G, Ioris RM, Villanyi Z, Firnkes R, Panasenko OO, Allen G, et al. Fkbp10 regulates protein translation to sustain lung cancer growth. Cell Rep. 2020;30(11):3851–63.
Madhav A, Andres A, Duong F, Mishra R, Haldar S, Liu Z, et al. Antagonizing cd105 enhances radiation sensitivity in prostate cancer. Oncogene. 2018;37(32):4385–97.
Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6(8):1259–67.
Ren X, Chen X, Fang K, Zhang X, Wei X, Zhang T, et al. Col5a2 promotes proliferation and invasion in prostate cancer and is one of seven gleason-related genes that predict recurrence-free survival. Front Oncol. 2021;11:583083.
Angel PM, Spruill L, Jefferson M, Bethard JR, Ball LE, Hughes-Halbert C, et al. Zonal regulation of collagen-type proteins and posttranslational modifications in prostatic benign and cancer tissues by imaging mass spectrometry. Prostate. 2020;80(13):1071–86.
Wang Z, Deng T, Long X, Lin X, Wu S, Wang H, et al. Methylation of srd5a2 promoter predicts a better outcome for castration-resistant prostate cancer patients undergoing androgen deprivation therapy. PLoS ONE. 2020;15(3): e229754.
Shiota M, Akamatsu S, Narita S, Sumiyoshi T, Fujiwara M, Uchiumi T, et al. The association between missense polymorphisms in srd5a2 and hsd3b1 and treatment failure with abiraterone for castration-resistant prostate cancer. Pharmacogenomics J. 2021;21(4):440–5.
Sun DY, Wu JQ, He ZH, He MF, Sun HB, et al. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through tgf-beta signaling pathway. Life Sci. 2019;235:116791.
Barcellos-De-Souza P, Comito G, Pons-Segura C, Taddei ML, Gori V, Becherucci V, et al. Mesenchymal stem cells are recruited and activated into carcinoma-associated fibroblasts by prostate cancer microenvironment-derived tgf-beta1. Stem Cells. 2016;34(10):2536–47.
Wu T, Wang W, Shi G, Hao M, Wang Y, Yao M, et al. Targeting hic1/tgf-beta axis-shaped prostate cancer microenvironment restrains its progression. Cell Death Dis. 2022;13(7):624.
Shi X, Young CD, Zhou H, Wang X, et al. Transforming growth factor-beta signaling in fibrotic diseases and cancer-associated fibroblasts. Biomolecules. 2020;10:12.
Ioannidou E, Moschetta M, Shah S, Parker JS, Ozturk MA, Pappas-Gogos G, et al. Angiogenesis and anti-angiogenic treatment in prostate cancer: mechanisms of action and molecular targets. Int J Mol Sci. 2021;22:18.
Zhang R, Liu F, et al. Cancer-associated fibroblast-derived gene signatures predict radiotherapeutic survival in prostate cancer patients. J Transl Med. 2022;20(1):453.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised recist guideline (version 1.1). Eur J Cancer. 2009;45(2):228–47.
Acknowledgements
We would like to thank TCGA and ENA databases for their contributions.
Funding
This study was supported by Key Areas Research and Development Program of Guangdong (2023B1111030006), National Natural Science Foundation of China (82372766 and 82072841), Natural Science Foundation of Guangdong Province (2021A1515010199), Key Areas Research and Development Program of Guangdong (2020B111114002), Guangdong Provincial Clinical Research Center for Urological Diseases (2020B1111170006) and Guangdong Science and Technology Department (2020B1212060018).
Author information
Authors and Affiliations
Contributions
CL: data curation, formal analysis, writing—original draft, writing—review and editing. ZW: data curation, formal analysis, writing—original draft, writing—review and editing. ZL: formal analysis, writing—original draft, writing—review and editing. XH: data curation, writing—original draft, writing—review and editing. ZH: formal analysis, writing—review and editing. HY: formal analysis, writing—review and editing. ZY: formal analysis, writing—original draft, writing—review and editing. JS: formal analysis, writing—review and editing. JH: writing—review and editing. YM: writing—review and editing. CL: conceptualization, funding acquisition, supervision, writing—review and editing. KX: conceptualization, funding acquisition, supervision, writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lai, C., Wu, Z., Li, Z. et al. Single-cell analysis extracted CAFs-related genes to established online app to predict clinical outcome and radiotherapy prognosis of prostate cancer. Clin Transl Oncol 26, 1240–1255 (2024). https://doi.org/10.1007/s12094-023-03348-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03348-6