Abstract
Purpose
Primary bone and joint sarcomas of the long bone are relatively rare neoplasms with poor prognosis. An efficient clinical tool that can accurately predict patient prognosis is not available. The current study aimed to use deep learning algorithms to develop a prediction model for the prognosis of patients with long bone sarcoma.
Methods
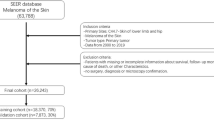
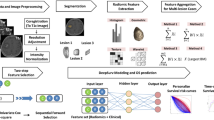
Data of patients with long bone sarcoma in the extremities was collected from the Surveillance, Epidemiology, and End Results Program database from 2004 to 2014. Univariate and multivariate analyses were performed to select possible prediction features. DeepSurv, a deep learning model, was constructed for predicting cancer-specific survival rates. In addition, the classical cox proportional hazards model was established for comparison. The predictive accuracy of our models was assessed using the C-index, Integrated Brier Score, receiver operating characteristic curve, and calibration curve.
Results
Age, tumor extension, histological grade, tumor size, surgery, and distant metastasis were associated with cancer-specific survival in patients with long bone sarcoma. According to loss function values, our models converged successfully and effectively learned the survival data of the training cohort. Based on the C-index, area under the curve, calibration curve, and Integrated Brier Score, the deep learning model was more accurate and flexible in predicting survival rates than the cox proportional hazards model.
Conclusion
A deep learning model for predicting the survival probability of patients with long bone sarcoma was constructed and validated. It is more accurate and flexible in predicting prognosis than the classical CoxPH model.






Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study. The datasets used and analyzed in the study are available from the corresponding author on reasonable request.
Abbreviations
- C-index:
-
Concordance index
- CSS:
-
Cancer-specific survival
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- SEER:
-
Surveillance, Epidemiology, and End Results
- IBS:
-
Integrated Brier Score
- ML:
-
Machine learning
- CoxPH:
-
Cox proportional hazards
- AI:
-
Artificial intelligence
References
Biermann JS, Adkins DR, Agulnik M, Benjamin RS, Brigman B, Butrynski JE, et al. Bone cancer. J Natl Compr Canc Netw. 2013;11(6):688–723. https://doi.org/10.6004/jnccn.2013.0088.
SEER. Cancer Stat Facts: Bone and Joint Cancer. National Cancer Institute, Bethesda. Available online: https://seer.cancer.gov/statfacts/html/bones.html. Accessed 5 May 2022.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA A Cancer J Clin. 2022;72(1):7–33. https://doi.org/10.3322/caac.21708.
Xu G, Wu H, Xu Y, Zhang Y, Lin F, Baklaushev VP, et al. Homogenous and heterogenous prognostic factors for patients with bone sarcoma. Orthop Surg. 2021;13(1):134–44. https://doi.org/10.1111/os.12851.
Hu X, Deng K, Ye H, Sun Z, Huang W, Sun Y, et al. Trends in tumor site-specific survival of bone sarcomas from 1980 to 2018: a surveillance, epidemiology and end results-based study. Cancers. 2021;13(21):5381. https://doi.org/10.3390/cancers13215381.
Biermann JS, Chow W, Reed DR, Lucas D, Adkins DR, Agulnik M, et al. NCCN guidelines insights: bone cancer, version 2.2017. J Natl Compr Canc Netw. 2017;15(2):155–67. https://doi.org/10.6004/jnccn.2017.0017.
Gospodarowicz M, Benedet L, Hutter RV, Fleming I, Henson DE, Sobin LH. History and international developments in cancer staging. Cancer Prev Control. 1998;2(6):262–8.
Heare T, Hensley MA, DellʼOrfano S. Bone tumors: osteosarcoma and Ewingʼs sarcoma. Curr Opin Pediatr. 2009;21(3):365–72. https://doi.org/10.1097/MOP.0b013e32832b1111.
Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–70. https://doi.org/10.1200/JCO.2007.12.9791.
Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–80. https://doi.org/10.1016/S1470-2045(14)71116-7.
Tian S, Liu S, Qing X, Lin H, Peng Y, Wang B, et al. A predictive model with a risk-classification system for cancer-specific survival in patients with primary osteosarcoma of long bone. Transl Oncol. 2022;18:101349. https://doi.org/10.1016/j.tranon.2022.101349.
Huang Y, Wang C, Tang D, Chen B, Jiang Z. Development and validation of nomogram-based prognosis tools for patients with extremity osteosarcoma: a SEER population study. J Oncol. 2022;2022:1–17. https://doi.org/10.1155/2022/9053663.
Wu X, Wang Y, Sun W, Tan M. Prognostic factors and a nomogram predicting overall survival in patients with limb chondrosarcomas: a population-based study. Biomed Res Int. 2021;2021:1–17. https://doi.org/10.1155/2021/4510423.
Fotso S. Deep Neural Networks for Survival Analysis Based on a Multi-Task Framework. arXiv: Mach Learn. 2018;arXiv:1801.05512.
Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020;471:61–71. https://doi.org/10.1016/j.canlet.2019.12.007.
Goecks J, Jalili V, Heiser LM, Gray JW. How machine learning will transform biomedicine. Cell. 2020;181(1):92–101. https://doi.org/10.1016/j.cell.2020.03.022.
Byun S, Heo TS, Choi JM, Jeong YS, Kim YS, Lee WK, et al. Deep learning based prediction of prognosis in nonmetastatic clear cell renal cell carcinoma. Sci Rep. 2021. https://doi.org/10.1038/s41598-020-80262-9.
Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018. https://doi.org/10.1186/s12874-018-0482-1.
Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–73. https://doi.org/10.1016/S1470-2045(19)30149-4.
Warren JL, Klabunde CN, Schrag D, Bach PB, Riley GF. Overview of the SEER-Medicare data: content, research applications, and generalizability to the United States elderly population. Med Care. 2002;40(8 Suppl):3–18. https://doi.org/10.1097/01.MLR.0000020942.47004.03.
Aran V, Devalle S, Meohas W, Heringer M, Cunha Caruso A, Pinheiro Aguiar D, et al. Osteosarcoma, chondrosarcoma and Ewing sarcoma: clinical aspects, biomarker discovery and liquid biopsy. Crit Rev Oncol Hematol. 2021;162:103340. https://doi.org/10.1016/j.critrevonc.2021.103340.
Stiller CA, Trama A, Serraino D, Rossi S, Navarro C, Chirlaque MD, et al. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur J Cancer. 2013;49(3):684–95. https://doi.org/10.1016/j.ejca.2012.09.011.
Ingley KM, Maleddu A, Grange FL, Gerrand C, Bleyer A, Yasmin E, et al. Current approaches to management of bone sarcoma in adolescent and young adult patients. Pediatr Blood Cancer. 2022. https://doi.org/10.1002/pbc.29442.
Strauss SJ, Frezza AM, Abecassis N, Bajpai J, Bauer S, Biagini R, et al. Bone sarcomas: ESMO–EURACAN–GENTURIS–ERN PaedCan clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol. 2021;32(12):1520–36. https://doi.org/10.1016/j.annonc.2021.08.1995.
Huang C, Su Q, Ding Z, Zeng W, Zhou Z. A novel clinical tool to predict cancer-specific survival in patients with primary pelvic sarcomas: a large population-based retrospective cohort study. Cancer Med-Us. 2022. https://doi.org/10.1002/cam4.4998.
Funding
This work was supported by National Natural Science Foundation of China (No. 31971272), Sanming Project of Medicine in Shenzhen (No. SZSM201911011) and International Science and Technology Cooperation Key Program project of Shaanxi Province (No.2023-GHZD-25).
Author information
Authors and Affiliations
Contributions
Data curation, ZM; Formal analysis, DC; Funding acquisition, HF; Investigation, XL, ZZ, JD, DZ; Methodology, DC, DL, ZZ, and JF; Project administration, HF; Resources, DC, ZM; Software, DC and WT; Supervision, HF; Validation, DL and XL; Visualization, HF; Writing–original draft, DC; Writing–review & editing, HF.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
Because the SEER database is a publicly available database of de-identified patient data, no ethics committee review was required for its use in this project.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, D., Liu, D., Li, X. et al. A deep learning model for accurately predicting cancer-specific survival in patients with primary bone sarcoma of the extremity: a population-based study. Clin Transl Oncol 26, 709–719 (2024). https://doi.org/10.1007/s12094-023-03291-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03291-6