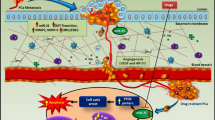
Abstract
Despite recent therapy advances and a better understanding of colon cancer biology, it remains one of the major causes of death. The cancer stem cells, associated with the progression, metastasis, and recurrence of colon cancer, play a major role in promoting the development of tumour and are found to be chemo resistant. The stroma of the tumour, which makes up the bulk of the tumour mass, is composed of the tumour microenvironment. With the advent of theranostic and the development of personalised medicine, miRNAs are becoming increasingly important in the context of colon malignancies. A holistic understanding of the regulatory roles of miRNAs in cancer cells and cancer stem cells will allow us to design effective strategies to regulate miRNAs, which could lead to improved clinical translation and creating a potent colon cancer treatment strategy. In this review paper, we briefly discuss the history of miRNA as well as the mechanisms of miRNA and cancer stem cells that contribute to the tumour growth, apoptosis, and advancement of colon cancer. The usefulness of miRNA in colorectal cancer theranostic is further concisely reviewed. We conclude by holding a stance in addressing the prospects and possibilities for miRNA by the disclosure of recent theranostic approaches aimed at eradicating cancer stem cells and enhancing overall cancer treatment outcomes.


Similar content being viewed by others
Data availability
NA.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Nusse R, Brown A, Papkoff J, Scambler P, Shackleford G, McMahon A, Moon R, et al. A new nomenclature for int-1 and related genes: the Wnt gene family. Cell. 1991;64:231. https://doi.org/10.1016/0092-8674(91)90633-a.
Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9:1–4. https://doi.org/10.1186/s13578-019-0361-4.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. https://doi.org/10.1038/35888.
Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, et al. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–68. https://doi.org/10.1093/nar/gkq727.
Forterre A, Komuro H, Aminova S, Harada M. A comprehensive review of cancer MicroRNA therapeutic delivery strategies. Cancers (Basel). 2020;12:1852. https://doi.org/10.3390/cancers12071852.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. https://doi.org/10.1101/gr.082701.108.
Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, et al. Fecal microRNAs as novel biomarkers for colon cancer screening fecal micrornas and cancer biomarkers. Cancer Epidemiol Biomark Prev. 2010;19:1766–74. https://doi.org/10.1158/1055-9965.EPI-10-0027.
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. https://doi.org/10.1073/pnas.1019055108.
Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199:407–12. https://doi.org/10.1083/jcb.201208082.
Paul S, Vazquez LA, Uribe SP, Cardenas LA, Aguilar MF, Chakraborty S, et al. Roles of microRNAs in carbohydrate and lipid metabolism disorders and their therapeutic potential. Biochimie. 2021;187:83–93. https://doi.org/10.1016/j.biochi.2021.05.015.
Wu X, Yan F, Wang L, Sun G, Liu J, Qu M, et al. MicroRNA: another pharmacological avenue for colorectal cancer? Front Cell Dev Biol. 2020;8:812. https://doi.org/10.3389/fcell.2020.00812.
Bravo V, Rosero S, Ricordi C, Pastori RL. Instability of miRNA and cDNAs derivatives in RNA preparations. Biochem Biophys Res Commun. 2007;353:1052–5. https://doi.org/10.1016/j.bbrc.2006.12.135.
Bernardo BC, Charchar FJ, Lin RC, McMullen JR. A microRNA guide for clinicians and basic scientists: background and experimental techniques. Heart Lung Circ. 2012;21:131–42. https://doi.org/10.1016/j.hlc.2011.11.002.
Khatri NI, Rathi MN, Baradia DP, Trehan S, Misra A. In vivo delivery aspects of miRNA, shRNA, and siRNA. Crit Rev Ther Drug Carrier Syst. 2012;29:487–527. https://doi.org/10.1615/critrevtherdrugcarriersyst.v29.i6.20.
Xie T, Li L. Stem cells and their niche: an inseparable relationship. Development. 2007;134:2001–6. https://doi.org/10.1242/dev.002022.
Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. https://doi.org/10.1083/jcb.201102147.
Wong GS, Rustgi AK. Matricellular proteins: priming the tumour microenvironment for cancer development and metastasis. Br J Cancer. 2013;108:755–61. https://doi.org/10.1038/bjc.2012.592.
Cuiffo BG, Karnoub AE. Mesenchymal stem cells in tumor development: emerging roles and concepts. Cell Adh Migr. 2012;6:220–30. https://doi.org/10.4161/cam.20875.
Nishimura K, Semba S, Aoyagi K, Sasaki H, Yokozaki H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology. 2012;79:290–306. https://doi.org/10.1159/000337296.
Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–7. https://doi.org/10.1016/j.coi.2010.01.009.
Jinushi M, Baghdadi M, Chiba S, Yoshiyama H. Regulation of cancer stem cell activities by tumor-associated macrophages. Am J Cancer Res. 2012;2:529–39.
Yu X, Li H, Ren X. Interaction between regulatory T cells and cancer stem cells. Int J Cancer. 2012;131:1491–8. https://doi.org/10.1002/ijc.27634.
Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–9. https://doi.org/10.1172/JCI57099.
Ye J, Wu D, Wu P, Chen Z, Huang J. The cancer stem cell niche: cross talk between cancer stem cells and their microenvironment. Tumour Biol. 2014;35:3945–51. https://doi.org/10.1007/s13277-013-1561-x.
Yu Y, Sarkar FH, Majumdar AP. Down-regulation of miR-21 induces differentiation of chemoresistant colon cancer cells and enhances susceptibility to therapeutic regimens. Transl Oncol. 2013;6:180–6. https://doi.org/10.1593/tlo.12397.
Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802. https://doi.org/10.1158/0008-5472.CAN-08-0951.
Ma Y, Zhang P, Wang F, Zhang H, Yang Y, Shi C, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1–2. https://doi.org/10.1038/ncomms2276.
Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12(5):602–15. https://doi.org/10.1016/j.stem.2013.03.002.
Cheng D, Zhao S, Tang H, Zhang D, Sun H, Yu F, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7:45199–213. https://doi.org/10.18632/oncotarget.9900.
Liu M, Xu A, Yuan X, Zhang Q, Fang T, Wang W, et al. Downregulation of microRNA-409-3p promotes aggressiveness and metastasis in colorectal cancer: an indication for personalized medicine. J Transl Med. 2015;13:1–9. https://doi.org/10.1186/s12967-015-0533-x.
Lu YX, Yuan L, Xue XL, Zhou M, Liu Y, Zhang C, et al. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2–negative feedback loop mechanism. Clin Cancer Res. 2014;20:2631–42. https://doi.org/10.1158/1078-0432.CCR-13-2348.
Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–71. https://doi.org/10.1002/stem.741.
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z, et al. MiRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS ONE. 2013;8:e60687. https://doi.org/10.1371/journal.pone.0060687.
Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–80. https://doi.org/10.1038/s41556-019-0286-5.
Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63.
Heijden MV, Vermeulen LA. Cancer stem cell perspective on minimal residual disease in solid malignancies incancer stem cell resistance to targeted therapy. Front Immunol. 2019;11:1280. https://doi.org/10.3389/fimmu.2020.01280.
Jahanafrooz Z, Mosafer J, Akbari M, Hashemzaei M, Mokhtarzadeh A, Baradaran B. Colon cancer therapy by focusing on colon cancer stem cells and their tumor microenvironment. J Cell Physiol. 2020;235:4153–66. https://doi.org/10.1002/jcp.29337.
De Robertis M, Poeta ML, Signori E, Fazio VM. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol. 2018;53:232–47. https://doi.org/10.1016/j.semcancer.2018.08.008.
Stark VA, Facey CO, Viswanathan V, Boman BM. The role of miRNAs, miRNA clusters, and isomiRs in development of cancer stem cell populations in colorectal cancer. Int J Mol Sci. 2021;22:1424. https://doi.org/10.3390/ijms22031424.
Shirmohamadi M, Eghbali E, Najjary S, Mokhtarzadeh A, Kojabad AB, Hajiasgharzadeh K, et al. Regulatory mechanisms of microRNAs in colorectal cancer and colorectal cancer stem cells. J Cell Physiol. 2020;235:776–89. https://doi.org/10.1002/jcp.29042.
Das PK, Islam F, Lam AK. The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells. 2020;9:1392. https://doi.org/10.3390/cells9061392.
Ullmann P, Nurmik M, Schmitz M, Rodriguez F, Weiler J, Qureshi-Baig K, et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019;450:32–41. https://doi.org/10.1016/j.canlet.2019.02.030.
Angius A, Scanu AM, Arru C, Muroni MR, Rallo V, Deiana G, et al. Portrait of cancer stem cells on colorectal cancer: Molecular biomarkers, signaling pathways and miRNAome. Int J Mol Sci. 2021;22:1603. https://doi.org/10.3390/ijms22041603.
Ye J, Lei J, Fang Q, Shen Y, Xia W, Hu X, et al. miR-4666-3p and miR-329 synergistically suppress the stemness of colorectal cancer cells via targeting TGF-β/Smad pathway. Front Oncol. 2019;9:1251. https://doi.org/10.3389/fonc.2019.01251.
Poturnajova M, Furielova T, Balintova S, Schmidtova S, Kucerova L, Matuskova M. Molecular features, and gene expression signature of metastatic colorectal cancer. Oncol Rep. 2012. https://doi.org/10.3892/or.2021.7961.
Mélin C, Perraud A, Akil H, Jauberteau MO, Cardot P, Mathonnet M, et al. Cancer stem cell sorting from colorectal cancer cell lines by sedimentation field flow fractionation. Anal Chem. 2012;84(3):1549–56. https://doi.org/10.1021/ac202797z.
Anderson EC, Hessman C, Levin TG, Monroe MM, Wong MH. The role of colorectal cancer stem cells in metastatic disease and therapeutic response. Cancers. 2011;3(1):319–39. https://doi.org/10.3390/cancers3010319.
Galizia G, Gemei M, Del Vecchio L, Zamboli A, Di Noto R, Mirabelli P, et al. Combined CD133/CD44 expression as a prognostic indicator of disease-free survival in patients with colorectal cancer. Arch Surg. 2012;147(1):18–24. https://doi.org/10.1001/archsurg.2011.795.
Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, et al. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111(2):251–63. https://doi.org/10.1016/S0092-8674(02)01015-2.
MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4(12):007880. https://doi.org/10.1101/cshperspect.a007880.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108(6):837–47. https://doi.org/10.1016/S0092-8674(02)00685-2.
Yan KS, Janda CY, Chang J, Zheng GX, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature. 2017;545(7653):238–42. https://doi.org/10.1038/nature22313.
Liu D, Du L, Chen D, Ye Z, Duan H, Tu T, et al. Reduced CD146 expression promotes tumorigenesis and cancer stemness in colorectal cancer through activating Wnt/β-catenin signaling. Oncotarget. 2016;7(26):40704. https://doi.org/10.18632/oncotarget.9930.
Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, et al. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33(6):1223–9. https://doi.org/10.3892/ijo_00000112.
Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–58. https://doi.org/10.1053/j.gastro.2008.02.057.
Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J Cell Mol Med. 2010;14(12):2697–701. https://doi.org/10.1111/j.1582-4934.2010.01191.x.
Li Q, Lex RK, Chung H, Giovanetti SM, Ji Z, Ji H, et al. The pluripotency factor NANOG binds to GLI proteins and represses hedgehog-mediated transcription. JBC. 2016;291(13):7171–82. https://doi.org/10.1074/jbc.M116.714857.
Oniscu A, James RM, Morris RG, Bader S, Malcomson RD, Harrison DJ. Expression of Sonic hedgehog pathway genes is altered in colonic neoplasia. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2004;203(4):909–17. https://doi.org/10.1002/path.1591.
Bian YH, Huang SH, Yang L, Ma XL, Xie JW, Zhang HW. Sonic hedgehog-Gli1 pathway in colorectal adenocarcinomas. WJG. 2009;13(11):1659.
Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1(6–7):338–51. https://doi.org/10.1002/emmm.200900039.
Wahab S, Alshahrani MY, Ahmad MF, Abbas H. Current trends, and future perspectives of nanomedicine for the management of colon cancer. Eur J Pharmacol. 2021;910:174464. https://doi.org/10.1016/j.ejphar.2021.174464.
Dembic Z. Antitumor drugs and their targets. Molecules. 2020;25(23):5776. https://doi.org/10.3390/molecules25235776.
Jothimani G, Bhatiya M, Pathak S, Paul S, Banerjee A. Tumor suppressor microRNAs in gastrointestinal cancers: A mini review. Recent Adv Inflammat Allergy Drug Discov. 2022. https://doi.org/10.2174/2772270816666220606112727.
Krajewska JB, Fichna J, Mosińska P. One step ahead: miRNA-34 in colon cancer-future diagnostic and therapeutic tool? Crit Rev Oncol Hematol. 2018;132:1–8. https://doi.org/10.1016/j.critrevonc.2018.09.006.
Li W, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, et al. MicroRNA-34a: potent tumor suppressor, cancer stem cell inhibitor, and potential anticancer therapeutic. Front Cell Dev Biol. 2021;9:640587. https://doi.org/10.3389/fcell.2021.640587.
Kim D, Kim Y, Kim Y. Effects of β-carotene on expression of selected microRNAs, histone acetylation, and DNA methylation in colon cancer stem cells. JCP. 2019;24(4):224.
Long J, He Q, Yin Y, Lei X, Li Z, Zhu W. The effect of miRNA and autophagy on colorectal cancer. Cell Prolif. 2020;53(10):e12900. https://doi.org/10.1111/cpr.12900.
Asadzadeh Z, Mansoori B, Mohammadi A, Aghajani M, Haji-Asgarzadeh K, Safarzadeh E, et al. MicroRNAs in cancer stem cells: Biology, pathways, and therapeutic opportunities. J Cell Physiol. 2019;234(7):10002–17. https://doi.org/10.1002/jcp.27885.
Das PK, Zahan T, Abdur Rakib M, Khanam JA, Pillai S, Islam F. Natural Compounds Targeting Cancer Stem Cells: A Promising Resource for Chemotherapy. Anticancer Agents Med Chem. 2019;19(15):1796–808. https://doi.org/10.2174/1871520619666190704111714.
Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY). 2016;8(4):603.
Garza Treviño EN, González PD, Valencia Salgado CI, Martinez GA. Effects of pericytes and colon cancer stem cells in the tumor microenvironment. Cancer Cell Int. 2019;19(1):1–2. https://doi.org/10.1186/s12935-019-0888-9.
Akshaya K, Arthi C, Pavithra AJ, Poovizhi P, Antinate SS, Hikku GS, et al. Bioconjugated gold nanoparticles as an efficient colorimetric sensor for cancer diagnostics. Photodiagn Photodyn Therapy. 2020;30:101699. https://doi.org/10.1016/j.pdpdt.2020.101699.
Girigoswami K, Girigoswami A. A review on the role of nanosensors in detecting cellular miRNA expression in colorectal cancer. EMIDDT. 2021;21(1):12–26. https://doi.org/10.2174/1871530320666200515115723.
Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies, and challenges. Nat Rev Drug Discov. 2010;9(10):775–89. https://doi.org/10.1038/nrd3179.
Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18(12):1121–6. https://doi.org/10.1038/gt.2011.79.
Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discovery Today. 2013;18(5–6):282–9. https://doi.org/10.1016/j.drudis.2012.10.002.
Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903–6. https://doi.org/10.1248/bpb.29.903.
Jaratlerdsiri W, Chan EK, Gong T, Petersen DC, Kalsbeek AM, Venter PA, et al. Whole-genome sequencing reveals elevated tumor mutational burden and initiating driver mutations in African men with treatment-naïve, high-risk prostate cancer. Cancer Res. 2018;78(24):6736–46. https://doi.org/10.1158/0008-5472.CAN-18-0254.
Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–80. https://doi.org/10.1038/nrd4278.
Bravo Vázquez LA, Moreno Becerril MY, Mora Hernández EO, León Carmona GG, Aguirre Padilla ME, Chakraborty S, et al. The emerging role of microRNAs in bone diseases and their therapeutic potential. Molecules. 2021;27(1):211. https://doi.org/10.3390/molecules27010211.
Ruiz-Manriquez LM, Estrada-Meza C, Benavides-Aguilar JA, Ledesma-Pacheco SJ, Torres-Copado A, Serrano-Cano FI, et al. Phytochemicals mediated modulation of microRNAs and long non-coding RNAs in cancer prevention and therapy. Phytother Res. 2022;36(2):705–29. https://doi.org/10.1002/ptr.7338.
Pandi S, Kulanthaivel L, Subbaraj GK, Rajaram S, Subramanian S. Screening of potential breast cancer inhibitors through molecular docking and molecular dynamics simulation. Biomed Res Int. 2022. https://doi.org/10.1155/2022/3338549.
Acknowledgements
The authors of this paper are thankful to Chettinad Academy of Research and Education (CARE) for providing the support infrastructurally and to DST, SERB, Govt of India, for the support.
Funding
This work was supported by the DST Inspire research student grant with award number 190963 and Departmental Seed Grants to Dr. Antara Banerjee from Chettinad academy of Research and Education with reference number -Ref.No.004/Regr/AR-Research/2022-05.
Author information
Authors and Affiliations
Contributions
The study was designed, and the manuscript was written by AB, DD, MK, CC, XS and reviewed and edited by SP, AB, AZ, HZ and SP.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that there are no conflicts of interests or competing interests.
Ethical approval and Informed consent
Ethical approval and informed consent are not applicable for the present work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Banerjee, A., Deka, D., Muralikumar, M. et al. A concise review on miRNAs as regulators of colon cancer stem cells and associated signalling pathways. Clin Transl Oncol 25, 3345–3356 (2023). https://doi.org/10.1007/s12094-023-03200-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03200-x