Abstract
Background
The lncRNA HOTAIR is frequently overexpressed in breast cancer tissues and plays an important role in the development of breast cancer. Here, we investigated the effect of the lncRNA HOTAIR on the biological behaviour of breast cancer cells and its molecular mechanism.
Methods
We investigated the level of HOTAIR in breast cancer and its clinical pathological characteristics by bioinformatic methods. Then, we evaluated the effects of HOTAIR and miRNA-1 expression on the biological behaviour of breast cancer cells by qPCR, Cell Counting Kit-8 (CCK-8) assay, clonogenic assays, Transwell assay and flow cytometry for cell proliferation, invasion migration and apoptosis, and cell cycle analysis. Finally, the target genes of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis were validated by luciferase reports.
Results
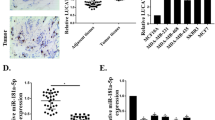
The expression of HOTAIR in breast cancer tissues was significantly higher than that in normal breast tissues (P < 0.05). Silencing of HOTAIR suppressed cell proliferation, invasion and migration, promoted apoptosis and induced G1 phase block in breast cancer (P < 0.0001). We also verified that miR-1 is a target of HOTAIR and that GOLPH3 is a target of miR-1 by luciferase reporter assays (P < 0.001).
Conclusions
The expression of HOTAIR was significantly elevated in breast cancer tissues. Reducing the expression of HOTAIR inhibited the proliferation, invasion and migration of breast cancer cells and promoted apoptosis, and the mechanism was mainly the effect of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis on the biological behaviour of breast cancer cells.




Similar content being viewed by others
Data availability
Data availability statement is available for this paper at https://doi.org/10.6084/m9.figshare.21506364.
Change history
02 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s12094-023-03224-3
References
World Health Organization. WHO mortality database. http://www.who.int/healthinfo/statistics/mortality_rawdata/en/. Published online 2020. Accessed 2022 Oct 01.
Sharifian A, Pourhoseingholi MA, Emadedin M, Nejad MR, Ashtari S, Hajizadeh N, et al. Burden of breast cancer in Iranian women is increasing. Asian Pac J Cancer Prev. 2015;16:5049–52.
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36.
Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:379.
Reiche K, Kasack K, Schreiber S, Luders T, Due E, Naume B, et al. Long non-coding RNAs differentially expressed between normal versus primary breast tumor tissues disclose converse changes to breast cancer-related protein-coding genes. PLoS ONE. 2014;9:e106076.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7.
Ramnarine VR, Kobelev M, Gibb EA, Nouri M, Lin D, Wang Y, et al. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur Urol. 2019;76:546–59.
Deng X, Li S, Kong F, Ruan H, Xu X, Zhang X, et al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics. 2020;10:265–80.
Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68.
Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–8.
Sumazin P, Yang X, Chiu H-S, Chung W-J, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81.
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83.
Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18.
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Alves C, Fonseca A, Muys B, Bueno R, Burger M, de Souza J, et al. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31:2827–32.
Guo J, McKenna SL, O’Dwyer ME, Cahill MR, O’Driscoll CM. RNA interference for multiple myeloma therapy: targeting signal transduction pathways. Expert Opin Ther Targets. 2016;20:107–21.
Bayram S, Sumbul A, Batmaci C, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumor Biol. 2015;36:3863–70.
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–100.
Hua Y, Duan S, Murmann AE, Larsen N, Kjems J, Lund AH, et al. miRConnect: identifying effector genes of miRNAs and miRNA families in cancer cells. PLoS ONE. 2011;6: e26521.
Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14.
Sorensen K, Thomassen M, Tan Q, Bak M, Cold S, Burton M, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529–36.
Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–55.
Wang Y, Gong G, Xu J, Zhang Y, Wu S, Wang S. Long noncoding RNA HOTAIR promotes breast cancer development by targeting ZEB1 via sponging miR-601. Cancer Cell Int. 2020;20:320.
Zhang M, Wu K, Zhang P, Qiu Y, Bai F, Chen H. HOTAIR facilitates endocrine resistance in breast cancer through ESR1/miR-130b-3p axis: comprehensive analysis of mRNA-miRNA-lncRNA network. Int J Gen Med. 2021;14:4653–63.
Wu C, Taylor R, Lane D, Ladinsky M, Weisz J, Howell K. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–75.
Buschman M, Rahajeng J, Field S. GOLPH3 links the Golgi, DNA damage, and cancer. Can Res. 2015;75:624–7.
Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, et al. Overexpression of GOLPH3 Promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012;18:4059–69.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81960542 and 81960517), Science and Technology Project of Yunnan Provincial Science and Technology Department (grant no. 202001AU070053, 202001AU070093 and 202201AY070001-169), Yunnan Health Training Project of High Level Talents (grant no. H-2019075) and Beijing Science and Technology Innovation Medical Development Foundation (grant no. KC2021-JK-0044-6).
Author information
Authors and Affiliations
Contributions
LZ and JWZ contributed to the experiments and clinical data interpretation. JWZ and SCT discussed the results and analysed the data and wrote the manuscript conceived and designed the experiments. YZZ, NW and XZ contributed to the statistical analysis. RG, HML and CXL contributed to polishing the English to improve the quality of this manuscript. SCT, KZ and DQL supervised and directed the overall project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised to include the co-corresponding authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Zhang, L., Zhao, Y. et al. Long noncoding RNA HOTAIR promotes breast cancer development through the lncRNA HOTAIR/miR-1/GOLPH3 axis. Clin Transl Oncol 25, 3420–3430 (2023). https://doi.org/10.1007/s12094-023-03197-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03197-3