Abstract
Background
Peritoneal metastasis (PM) is an important factor contributing to poor prognosis in patients with gastric cancer (GC). Transcriptomic sequencing has been used to explore the molecular changes in metastatic cancers, but comparing the bulk RNA-sequencing data between primary tumors and metastases in PM studies is unreasonable due to the small proportion of tumor cells in PM tissues.
Methods
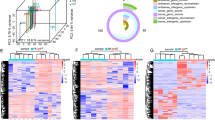
We performed single-cell RNA-sequencing analysis on four gastric adenocarcinoma specimens, including one primary tumor sample (PT), one adjacent nontumoral sample (PN), one peritoneal metastatic sample (MT) and one normal peritoneum sample (MN), from the same patient. Pseudotime trajectory analysis was used to display the process by which nonmalignant epithelial cells transform into tumor cells and then metastasize to the peritoneum. Finally, in vitro and in vivo assays were used to validate one of the selected genes that promote peritoneal metastasis.
Results
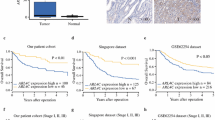
Single-cell RNA sequencing showed that a development curve was found from normal mucosa to tumor tissues and then into metastatic sites on peritoneum. TAGLN2 was found to trigger this metastasis process. The migration and invasion capability of GC cells were changed by downregulating and upregulating TAGLN2 expression. Mechanistically, TAGLN2 might modulate tumor metastasis via alterations in cell morphology and several signaling pathways, thus promoting epithelial–mesenchymal transition (EMT).
Conclusions
In summary, we identified and validated TAGLN2 as a novel gene involved in GC peritoneal metastasis. This study provided valuable insight into the mechanisms of GC metastasis and developed a potential therapeutic target to prevent GC cell dissemination.






Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author.
Change history
16 March 2024
A Correction to this paper has been published: https://doi.org/10.1007/s12094-024-03402-x
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Al-Batran SE, Homann N, Pauligk C, Illerhaus G, Martens UM, Stoehlmacher J, et al. Effect of neoadjuvant chemotherapy followed by surgical resection on survival in patients with limited metastatic gastric or gastroesophageal junction cancer: the AIO-FLOT3 trial. JAMA Oncol. 2017;3(9):1237–44. https://doi.org/10.1001/jamaoncol.2017.0515.
Koemans WJ, Lurvink RJ, Grootscholten C, Verhoeven RHA, de Hingh IH, van Sandick JW. Synchronous peritoneal metastases of gastric cancer origin: incidence, treatment and survival of a nationwide Dutch cohort. Gastric Cancer. 2021;24(4):800–9. https://doi.org/10.1007/s10120-021-01160-1.
Tan HL, Chia CS, Tan GH, Choo SP, Tai DW, Chua CW, et al. Gastric peritoneal carcinomatosis - a retrospective review. World J Gastrointest Oncol. 2017;9(3):121–8. https://doi.org/10.4251/wjgo.v9.i3.121.
Zhu M, Zhang N, He S, Lu X. Exosomal miR-106a derived from gastric cancer promotes peritoneal metastasis via direct regulation of Smad7. Cell Cycle. 2020;19(10):1200–21. https://doi.org/10.1080/15384101.2020.1749467.
Gwee YX, Chia DKA, So J, Ceelen W, Yong WP, Tan P, et al. Integration of genomic biology into therapeutic strategies of gastric cancer peritoneal metastasis. J Clin Oncol. 2022;40(24):2830. https://doi.org/10.1200/JCO.21.02745.
Nicolson GL, Belloni PN, Tressler RJ, Dulski K, Inoue T, Cavanaugh PG. Adhesive, invasive, and growth properties of selected metastatic variants of a murine large-cell lymphoma. Invasion Metastasis. 1989;9(2):102–16.
Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res. 2019;79(12):3011–27. https://doi.org/10.1158/0008-5472.CAN-19-0458.
Jo S, Kim HR, Mun Y, Jun CD. Transgelin-2 in immunity: its implication in cell therapy. J Leukoc Biol. 2018;104(5):903–10. https://doi.org/10.1002/JLB.MR1117-470R.
Sokolowska I, Dorobantu C, Woods AG, Macovei A, Branza-Nichita N, Darie CC. Proteomic analysis of plasma membranes isolated from undifferentiated and differentiated HepaRG cells. Proteome Sci. 2012;10(1):47. https://doi.org/10.1186/1477-5956-10-47.
Yulis M, Kusters DHM, Nusrat A. Cadherins: cellular adhesive molecules serving as signalling mediators. J Physiol. 2018;596(17):3883–98. https://doi.org/10.1113/JP275328.
Zhang JC, Helmke BP, Shum A, Du K, Yu WW, Lu MM, et al. SM22beta encodes a lineage-restricted cytoskeletal protein with a unique developmentally regulated pattern of expression. Mech Dev. 2002;115(1–2):161–6. https://doi.org/10.1016/s0925-4773(02)00088-6.
Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, et al. Role of transgelin-2 in diabetes-associated pancreatic ductal adenocarcinoma. Oncotarget. 2017;8(30):49592–604. https://doi.org/10.18632/oncotarget.17519.
Zhang Y, Ye Y, Shen D, Jiang K, Zhang H, Sun W, et al. Identification of transgelin-2 as a biomarker of colorectal cancer by laser capture microdissection and quantitative proteome analysis. Cancer Sci. 2010;101(2):523–9. https://doi.org/10.1111/j.1349-7006.2009.01424.x.
Jin H, Cheng X, Pei Y, Fu J, Lyu Z, Peng H, et al. Identification and verification of transgelin-2 as a potential biomarker of tumor-derived lung-cancer endothelial cells by comparative proteomics. J Proteomics. 2016;136:77–88. https://doi.org/10.1016/j.jprot.2015.12.012.
Shi YY, Wang HC, Yin YH, Sun WS, Li Y, Zhang CQ, et al. Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br J Cancer. 2005;92(5):929–34. https://doi.org/10.1038/sj.bjc.6602460.
Belczacka I, Latosinska A, Metzger J, Marx D, Vlahou A, Mischak H, et al. Proteomics biomarkers for solid tumors: Current status and future prospects. Mass Spectrom Rev. 2019;38(1):49–78. https://doi.org/10.1002/mas.21572.
Kim IG, Lee JH, Kim SY, Hwang HM, Kim TR, Cho EW. Hypoxia-inducible transgelin 2 selects epithelial-to-mesenchymal transition and gamma-radiation-resistant subtypes by focal adhesion kinase-associated insulin-like growth factor 1 receptor activation in non-small-cell lung cancer cells. Cancer Sci. 2018;109(11):3519–31. https://doi.org/10.1111/cas.13791.
Yin LM, Ulloa L, Yang YQ. Transgelin-2: biochemical and clinical implications in cancer and asthma. Trends Biochem Sci. 2019;44(10):885–96. https://doi.org/10.1016/j.tibs.2019.05.004.
Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12(3):670–91. https://doi.org/10.1158/2159-8290.CD-21-0683.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329-337 e324. https://doi.org/10.1016/j.cels.2019.03.003.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e1821. https://doi.org/10.1016/j.cell.2019.05.031.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14. https://doi.org/10.1016/j.cell.2015.05.002.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72. https://doi.org/10.1038/s41590-018-0276-y.
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, et al. Cell marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–8. https://doi.org/10.1093/nar/gky900.
Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–82. https://doi.org/10.1038/nmeth.4402.
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. https://doi.org/10.1038/s41586-019-0969-x.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. https://doi.org/10.1126/science.1254257.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. https://doi.org/10.1093/nar/gkx247.
Li H, Zhao J, Sun J, Tian C, Jiang Q, Ding C, et al. Demethylation of the SFRP4 promoter drives gastric cancer progression via the Wnt pathway. Mol Cancer Res. 2021;19(9):1454–64. https://doi.org/10.1158/1541-7786.MCR-20-0933.
Chothia C, Jones EY. The molecular structure of cell adhesion molecules. Annu Rev Biochem. 1997;66:823–62. https://doi.org/10.1146/annurev.biochem.66.1.823.
Fujitani K, Yang HK, Mizusawa J, Kim YW, Terashima M, Han SU, et al. Gastrectomy plus chemotherapy versus chemotherapy alone for advanced gastric cancer with a single non-curable factor (REGATTA): a phase 3, randomised controlled trial. Lancet Oncol. 2016;17(3):309–18. https://doi.org/10.1016/S1470-2045(15)00553-7.
Shimura T, Toden S, Kandimalla R, Toiyama Y, Okugawa Y, Kanda M, et al. Genomewide expression profiling identifies a novel miRNA-based signature for the detection of peritoneal metastasis in patients with gastric cancer. Ann Surg. 2021;274(5):e425–34. https://doi.org/10.1097/SLA.0000000000003647.
Li H, Fu X, Zhao J, Li C, Li L, Xia P, et al. EXOC4 promotes diffuse-type gastric cancer metastasis via activating FAK signal. Mol Cancer Res. 2022;20(7):1021–34. https://doi.org/10.1158/1541-7786.MCR-21-0441.
Bae H, Kim H, Chu J, Jang Y, Koh HH, Jung H, et al. Pathologic analyses of peritoneal nodules in gastric cancer patients during surgery-A single cancer center experience with diagnostic pitfalls. Pathol Res Pract. 2019;215(1):195–9. https://doi.org/10.1016/j.prp.2018.11.013.
Wang R, Dang M, Harada K, Han G, Wang F, Pool Pizzi M, et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat Med. 2021;27(1):141–51. https://doi.org/10.1038/s41591-020-1125-8.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. https://doi.org/10.1038/nature13480.
Wang R, Song S, Harada K, Ghazanfari Amlashi F, Badgwell B, Pizzi MP, et al. Multiplex profiling of peritoneal metastases from gastric adenocarcinoma identified novel targets and molecular subtypes that predict treatment response. Gut. 2020;69(1):18–31. https://doi.org/10.1136/gutjnl-2018-318070.
Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46(6):583–7. https://doi.org/10.1038/ng.2984.
Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. https://doi.org/10.1126/science.123.3191.309.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. https://doi.org/10.1016/j.ccr.2012.02.014.
LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. https://doi.org/10.1038/ncb3039.
Sun Y, Peng W, He W, Luo M, Chang G, Shen J, et al. Transgelin-2 is a novel target of KRAS-ERK signaling involved in the development of pancreatic cancer. J Exp Clin Cancer Res. 2018;37(1):166. https://doi.org/10.1186/s13046-018-0818-z.
Wang L, Tan H, Huang Y, Guo M, Dong Y, Liu C, et al. TAGLN2 promotes papillary thyroid carcinoma invasion via the Rap1/PI3K/AKT axis. Endocr Relat Cancer. 2023. https://doi.org/10.1530/ERC-21-0352.
Liu L, Wu N, Wang Y, Zhang X, Xia B, Tang J, et al. TRPM7 promotes the epithelial-mesenchymal transition in ovarian cancer through the calcium-related PI3K/AKT oncogenic signaling. J Exp Clin Cancer Res. 2019;38(1):106. https://doi.org/10.1186/s13046-019-1061-y.
Sheng W, Shi X, Lin Y, Tang J, Jia C, Cao R, et al. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin Cancer Res. 2020;39(1):16. https://doi.org/10.1186/s13046-020-1521-4.
Acknowledgements
This work was supported by grants awarded to Dr. Junjie Zhao (82002527), Prof. Xuefei Wang (81972228) and Prof. Yihong Sun (81872425) from National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
Conceptualization: CJ and JZ; methodology: CJ, HC, JZ. Data curation: CJ, CT, DL, CW; software and formal analysis: HC and CT; resources: TC, BY and MF; investigation: TC and JS; writing—original draft: CJ, JZ and ZW; writing—review and editing: XW and YS. Supervision, funding acquisition and project administration: HL and YS; all the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Ethical approval
All protocols in this study were approved by ethics committee of The Zhongshan Hospital and conducted in compliance with the Declaration of Helsinki.
Informed consent
Single-cell RNA sequencing data were used from public GEO datasets. All the tissues from patients were obtained after informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised to correct Fig 4E caption and Figure 5.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ji, C., Zhao, J., chen, H. et al. Single-cell RNA sequencing reveals the lineage of malignant epithelial cells and upregulation of TAGLN2 promotes peritoneal metastasis in gastric cancer. Clin Transl Oncol 25, 3405–3419 (2023). https://doi.org/10.1007/s12094-023-03194-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03194-6