Abstract
Background
Colon cancer with high incidence and mortality is a severe public health problem. As an emerging therapy, immunotherapy has played an active clinical role in tumor treatment, but only a small number of patients respond.
Methods
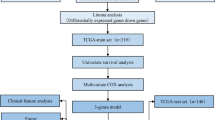
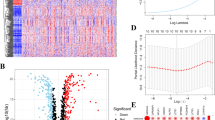
By univariate Cox regression analysis of 165 novel cancer prediction genes (NCPGs), 29 NCPGs related to prognosis were screened. Based on these 29 NCPGs and 336 differentially expressed genes, we constructed two colon cancer subgroups and three gene clusters and analyzed prognosis, activation pathways, and immune infiltration characteristics under various modification patterns. Then each patient was scored and divided into high or low NCPG_score groups. A comprehensive evaluation between NCPG_score and clinical characteristics, tumor microenvironment (TME), tumor somatic mutations, and the potential for immunotherapy was conducted.
Results
Patients with high NCPG_score were characterized by high tumor mutation burden and high microsatellite instability and were more suitable for immunotherapy.
Conclusions
This study screened 29 NCPGs as independent prognostic markers in colon cancer patients, demonstrating their TME, clinicopathological features, and potential roles in immunotherapy, helping to assess prognosis and guiding more personalized immunotherapy.






Similar content being viewed by others
Availability of data and materials
The data sets analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/), accession numbers TCGA-COAD, COAD-FPKM; GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39582).
Abbreviations
- CNV:
-
Copy number variation
- DEGs:
-
Differentially expressed genes
- FDA:
-
Food and Drug Administration
- ICB:
-
Immune-checkpoint blocker
- MSI:
-
Microsatellite instability
- MSI-H:
-
High microsatellite instability
- MDSCs:
-
Myeloid-derived suppressor cells
- NCPGs:
-
Novel cancer prediction genes
- PCA:
-
Principal component analysis
- ssGSEA:
-
Single sample gene set enrichment analysis
- TCGA:
-
The cancer genome Atlas
- TGF-beta:
-
Transforming growth factor beta
- TMB:
-
Tumor mutation burden
- TME:
-
Tumor microenvironment
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80.
Murphy N, Ward HA, Jenab M, Rothwell JA, Boutron-Ruault MC, Carbonnel F, et al. Heterogeneity of colorectal cancer risk factors by anatomical subsite in 10 European countries: a multinational cohort study. Clin Gastroenterol Hepatol. 2019;17(1323–31): e6.
Wong MCS, Huang J, Lok V, Wang J, Fung F, Ding H, et al. Differences in incidence and mortality trends of colorectal cancer worldwide based on sex, age, and anatomic location. Clin Gastroenterol Hepatol. 2021;19(955–66): e61.
Kim EK, Song MJ, Jung Y, Lee WS, Jang HH. Proteomic analysis of primary colon cancer and synchronous solitary liver metastasis. Cancer Genomics Proteomics. 2019;16:583–92.
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4.
Lee AY, Brady MS. Neoadjuvant immunotherapy for melanoma. J Surg Oncol. 2021;123:782–8.
Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. 2016;44:1255–69.
Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50.
Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, et al. T25 repeat in the 3’ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res. 2005;65:8072–8.
Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, inflammation and colorectal cancer. Cells. 2020;9:618.
Sahin IH, Akce M, Alese O, Shaib W, Lesinski GB, El-Rayes B, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121:809–18.
Wu Y, Xu J, Du C, Wu Y, Xia D, Lv W, et al. The predictive value of tumor mutation burden on efficacy of immune checkpoint inhibitors in cancers: a systematic review and meta-analysis. Front Oncol. 2019;9:1161.
Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096–103.
Wang X, Duanmu J, Fu X, Li T, Jiang Q. Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment. J Transl Med. 2020;18:1–14.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20.
Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9:5361.
Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35.
Singh MP, Rai S, Suyal S, Singh SK, Singh NK, Agarwal A, et al. Genetic and epigenetic markers in colorectal cancer screening: recent advances. Expert Rev Mol Diagn. 2017;17:665–85.
Zhou R, Zhang J, Zeng D, Sun H, Rong X, Shi M, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother. 2019;68:433–42.
Ju HQ, Zhao Q, Wang F, Lan P, Wang Z, Zuo ZX, et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol Med. 2019;11: e10168.
Wu Z, Lu Z, Li L, Ma M, Long F, Wu R, et al. Identification and validation of ferroptosis-related LncRNA signatures as a novel prognostic model for colon cancer. Front Immunol. 2021;12: 783362.
Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6.
Li X, Wen D, Li X, Yao C, Chong W, Chen H. Identification of an immune signature predicting prognosis risk and lymphocyte infiltration in colon cancer. Front Immunol. 2020;11:1678.
Luo D, Liu Q, Shan Z, Cai S, Li Q, Li X. Development and validation of a novel epigenetic signature for predicting prognosis in colon cancer. J Cell Physiol. 2020;235:8714–23.
Dai W, Li Y, Mo S, Feng Y, Zhang L, Xu Y, et al. A robust gene signature for the prediction of early relapse in stage I-III colon cancer. Mol Oncol. 2018;12:463–75.
Schulte-Sasse R, Budach S, Hnisz D, Marsico A. Integration of multiomics data with graph convolutional networks to identify new cancer genes and their associated molecular mechanisms. Nat Mach Intell. 2021;3:513–26.
Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49.
Shida D, Kanemitsu Y, Hamaguchi T, Shimada Y. Introducing the eighth edition of the tumor-node-metastasis classification as relevant to colorectal cancer, anal cancer and appendiceal cancer: a comparison study with the seventh edition of the tumor-node-metastasis and the Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma. Jpn J Clin Oncol. 2019;49:321–8.
Chin CC, Wang JY, Changchien CR, Huang WS, Tang R. Carcinoma obstruction of the proximal colon cancer and long-term prognosis–obstruction is a predictor of worse outcome in TNM stage II tumor. Int J Colorectal Dis. 2010;25:817–22.
Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–72.
Fan G, Lou L, Song Z, Zhang X, Xiong XF. Targeting mutated GTPase KRAS in tumor therapies. Eur J Med Chem. 2021;226: 113816.
McCuaig S, Barras D, Mann EH, Friedrich M, Bullers SJ, Janney A, et al. The interleukin 22 pathway interacts with mutant KRAS to promote poor prognosis in colon cancer. Clin Cancer Res. 2020;26:4313–25.
Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, et al. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13:12153–68.
Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–7.
Li Z, Jia Y, Zhu H, Xing X, Pang F, Shan F, et al. Tumor mutation burden is correlated with response and prognosis in microsatellite-stable (MSS) gastric cancer patients undergoing neoadjuvant chemotherapy. Gastric Cancer. 2021;24:1342–54.
Toh JWT, Mahajan H, Chapuis P, Spring K. Current status on microsatellite instability, prognosis and adjuvant therapy in colon cancer: a nationwide survey of medical oncologists, colorectal surgeons and gastrointestinal pathologists. Cancer Rep (Hoboken). 2021;4: e1297.
Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–21.
Stevens D, Ingels J, Van Lint S, Vandekerckhove B, Vermaelen K. Dendritic cell-based immunotherapy in lung cancer. Front Immunol. 2020;11: 620374.
Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E, et al. Immunotherapy for pancreatic cancer: a 2020 update. Cancer Treat Rev. 2020;86: 102016.
Henegan JC, Sonpavde G. Promising immunotherapy for prostate cancer. Expert Opin Biol Ther. 2018;18:109–20.
Hammers H. Immunotherapy in kidney cancer: the past, present, and future. Curr Opin Urol. 2016;26:543–7.
Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33:2780–8.
Basile D, Garattini SK, Bonotto M, Ongaro E, Casagrande M, Cattaneo M, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther. 2017;17:709–21.
Nakamura K, Smyth MJ. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol Immunol. 2020;17:1–12.
Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. 2019;120:16–25.
Pan PY, Ma G, Weber KJ, Ozao-Choy J, Wang G, Yin B, et al. Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res. 2010;70:99–108.
Shafabakhsh R, Pourhanifeh MH, Mirzaei HR, Sahebkar A, Asemi Z, Mirzaei H. Targeting regulatory T cells by curcumin: a potential for cancer immunotherapy. Pharmacol Res. 2019;147: 104353.
Derynck R, Turley SJ, Akhurst RJ. TGFbeta biology in cancer progression and immunotherapy. Nat Rev Clin Oncol. 2021;18:9–34.
Acknowledgements
The authors gratefully acknowledge TCGA and GEO databases providing their platforms for offering convenient access to datasets and the contributors for uploading their meaningful datasets.
Funding
This study was funded by the National Science Foundation of China (No. 81903539).
Author information
Authors and Affiliations
Contributions
SZ was responsible for the concept and design of the original study and performed a systematic search in the literature of original articles. SW performed the data analysis and manuscript preparation, SZ was responsible for the critical review of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Zhu, S. Comprehensive analysis of novel cancer prediction genes and tumor microenvironment infiltration in colon cancer. Clin Transl Oncol 25, 2545–2558 (2023). https://doi.org/10.1007/s12094-023-03145-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03145-1