Abstract
Background
Globally, gastric cancer (GC) is a common and lethal solid malignant tumor. Identifying the molecular signature and its functions can provide mechanistic insights into GC development and new methods for targeted therapy.
Methods
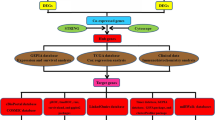
Differentially expressed genes (DEGs) and prognostic genes (from univariate Cox regression analysis) were overlapped to obtain prognostic DEGs. Subsequently, molecular modules and the functions of these prognostic DEGs were identified by Metascape and Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG)/Gene Set Enrichment Analysis (GSEA) enrichment analyses, respectively. Protein–protein interaction (PPI) networks of up- and down-regulated prognostic DEGs in GC were analyzed using the MCC algorithm of the Cytohubba plug-in in Cytoscape. The prognostic gene signature was defined on hub genes of the PPI networks by least absolute shrinkage and selection operator (LASSO)-Cox regression analysis. Furthermore, the expressional level of PLG in our clinical GC samples was validated by quantitative PCR (qPCR), western blotting, and immunohistochemistry (IHC). Subsequently, the PLG expression-correlation analysis was performed to assess the role of PLG in GC progression. Immune infiltration analysis was performed by single-sample gene set enrichment analysis (ssGSEA) to assess the inhibitory effect of PLG on immune infiltration.
Results
Firstly, Up- and down-regulated prognostic DEGs and hub genes in protein–protein interaction (PPI) networks in GC were identified. A prognostic five-gene signature (i.e., PLG, SPARC, FGB, SERPINE1, and KLHL41) was identified. Among the five genes, the relationship between plasminogen (PLG) and GC remains largely unclear. Moreover, the functions of PLG-correlated genes in GC, like 'fibrinolysis', 'hemostasis', 'ion channel complex', and 'transporter complex' were identified. In addition, PLG expression correlated negatively with the infiltration of almost all immune cell types. Interestingly, the expression of PLG was significantly and highly correlated with that of CD160, an immune checkpoint inhibitor.
Conclusion
Our findings defined a new five-gene signature for predicting GC prognosis, but more validation is required to assess the effects and mechanism of the five genes, especially PLG, for the development of new GC therapies.






Similar content being viewed by others
Availability of data and materials
Publicly available datasets were analyzed in this study. The data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi (GEO datasets), https://xenabrowser.net/datapages/ (TCGA datasets).
Abbreviations
- aDC:
-
Activated dendritic cells
- cDC:
-
Conventional dendritic cells
- iDC:
-
Immature dendritic cells
- pDC:
-
Plasmacytoid dendritic cells
- Tcm:
-
Central memory T-cells
- Tem:
-
Effector memory T cells
- Tgd:
-
T cells gamma delta
- TFH:
-
T follicular helper cells
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- GBM:
-
Glioblastoma multiforme
- STAD:
-
Stomach adenocarcinoma
- KICH:
-
Kidney chromophobe renal cell carcinoma
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- STAD:
-
Stomach adenocarcinoma
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. https://doi.org/10.3322/caac.21654.
Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626. https://doi.org/10.1177/1010428317714626.
Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020. https://doi.org/10.3390/ijms21114012.
Zhou L, Lu H, Zeng F, Zhou Q, Li S, Wu Y, et al. Constructing a new prognostic signature of gastric cancer based on multiple data sets. Bioengineered. 2021;12:2820–35. https://doi.org/10.1080/21655979.2021.1940030.
Lu XQ, Zhang JQ, Zhang SX, Qiao J, Qiu MT, Liu XR, et al. Identification of novel hub genes associated with gastric cancer using integrated bioinformatics analysis. BMC Cancer. 2021;21:697. https://doi.org/10.1186/s12885-021-08358-7.
Xie L, Cai L, Wang F, Zhang L, Wang Q, Guo X. Systematic review of prognostic gene signature in gastric cancer patients. Front Bioeng Biotechnol. 2020;8:805. https://doi.org/10.3389/fbioe.2020.00805.
Liu Q, Jiang J, Zhang X, Zhang M, Fu Y. Comprehensive analysis of IGFBPs as biomarkers in gastric cancer. Front Oncol. 2021;11: 723131. https://doi.org/10.3389/fonc.2021.723131.
Ren F, Zhao Q, Zhao M, Zhu S, Liu B, Bukhari I, et al. Immune infiltration profiling in gastric cancer and their clinical implications. Cancer Sci. 2021;112:3569–84. https://doi.org/10.1111/cas.15057.
Zhang AZ, Yuan X, Liang WH, Zhang HJ, Li Y, Xie YF, et al. Immune infiltration in gastric cancer microenvironment and its clinical significance. Front Cell Dev Biol. 2021;9: 762029. https://doi.org/10.3389/fcell.2021.762029.
Seillier C, Hélie P, Petit G, Vivien D, Clemente D, Le Mauff B, et al. Roles of the tissue-type plasminogen activator in immune response. Cell Immunol. 2022;371: 104451. https://doi.org/10.1016/j.cellimm.2021.104451.
Baker SK, Strickland S. A critical role for plasminogen in inflammation. J Exp Med. 2020. https://doi.org/10.1084/jem.20191865.
Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the urokinase-type plasminogen activator (uPA) and Its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018;8:24. https://doi.org/10.3389/fonc.2018.00024.
Wojtukiewicz MZ, Sierko E, Zacharski LR, Zimnoch L, Kudryk B, Kisiel W. Tissue factor-dependent coagulation activation and impaired fibrinolysis in situ in gastric cancer. Semin Thromb Hemost. 2003;29:291–300. https://doi.org/10.1055/s-2003-40967.
Ding Y, Zhang H, Lu A, Zhou Z, Zhong M, Shen D, et al. Effect of urokinase-type plasminogen activator system in gastric cancer with peritoneal metastasis. Oncol Lett. 2016;11:4208–16. https://doi.org/10.3892/ol.2016.4498.
Kit O, Frantsiyants E, Kozlova L, Maslov A, Kolesnikov E, Dzhenkova E, et al. Varying distribution of tissue plasminogen activators in gastrointestinal adenocarcinoma. Ann Oncol. 2018;29:v7. https://doi.org/10.1093/annonc/mdy151.023.
Plebani M, Herszènyi L, Carraro P, De Paoli M, Roveroni G, Cardin R, et al. Urokinase-type plasminogen activator receptor in gastric cancer: tissue expression and prognostic role. Clin Exp Metastasis. 1997;15:418–25. https://doi.org/10.1023/a:1018454305889.
Kaneko T, Konno H, Baba M, Tanaka T, Nakamura S. Urokinase-type plasminogen activator expression correlates with tumor angiogenesis and poor outcome in gastric cancer. Cancer Sci. 2003;94:43–9. https://doi.org/10.1111/j.1349-7006.2003.tb01350.x.
Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314–6. https://doi.org/10.1038/nbt.3772.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60. https://doi.org/10.1093/nar/gkz430.
Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020. https://doi.org/10.1126/science.aay5947.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. https://doi.org/10.1186/s13059-014-0550-8.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. https://doi.org/10.1038/75556.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. https://doi.org/10.1093/nar/28.1.27.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. https://doi.org/10.1089/omi.2011.0118.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. https://doi.org/10.1093/nar/gky1131.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-S4-S11.
Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. https://doi.org/10.18637/jss.v039.i05.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. https://doi.org/10.1186/1471-2105-12-77.
Li J, Shi H, Yuan Z, Wu Z, Li H, Liu Y, et al. The role of SPI1-TYROBP-FCER1G network in oncogenesis and prognosis of osteosarcoma, and its association with immune infiltration. BMC Cancer. 2022;22:108. https://doi.org/10.1186/s12885-022-09216-w.
Cheong JH, Wang SC, Park S, Porembka MR, Christie AL, Kim H, et al. Development and validation of a prognostic and predictive 32-gene signature for gastric cancer. Nat Commun. 2022;13:774. https://doi.org/10.1038/s41467-022-28437-y.
Wang J, Gao P, Song Y, Sun J, Chen X, Yu H, et al. Prognostic value of gastric cancer-associated gene signatures: evidence based on a meta-analysis using integrated bioinformatics methods. J Cell Mol Med. 2018;22:5743–7. https://doi.org/10.1111/jcmm.13823.
Zhao ZS, Wang YY, Chu YQ, Ye ZY, Tao HQ. SPARC is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res. 2010;16:260–8. https://doi.org/10.1158/1078-0432.CCR-09-1247.
Liao P, Li W, Liu R, Teer JK, Xu B, Zhang W, et al. Genome-scale analysis identifies SERPINE1 and SPARC as diagnostic and prognostic biomarkers in gastric cancer. Onco Targets Ther. 2018;11:6969–80. https://doi.org/10.2147/OTT.S173934.
Xu B, Bai Z, Yin J, Zhang Z. Global transcriptomic analysis identifies SERPINE1 as a prognostic biomarker associated with epithelial-to-mesenchymal transition in gastric cancer. PeerJ. 2019;7: e7091. https://doi.org/10.7717/peerj.7091.
Ren Q, Zhu P, Zhang H, Ye T, Liu D, Gong Z, et al. Identification and validation of stromal-tumor microenvironment-based subtypes tightly associated with PD-1/PD-L1 immunotherapy and outcomes in patients with gastric cancer. Cancer Cell Int. 2020;20:92. https://doi.org/10.1186/s12935-020-01173-3.
Yu X, Hu F, Yao Q, Li C, Zhang H, Xue Y. Serum fibrinogen levels are positively correlated with advanced tumor stage and poor survival in patients with gastric cancer undergoing gastrectomy: a large cohort retrospective study. BMC Cancer. 2016;16:480. https://doi.org/10.1186/s12885-016-2510-z.
Wijethilake N, Islam M, Ren H. Radiogenomics model for overall survival prediction of glioblastoma. Med Biol Eng Comput. 2020;58:1767–77. https://doi.org/10.1007/s11517-020-02179-9.
Choi SH, Cho SY, Song J, Hur MW. KLHL4, a novel p53 target gene, inhibits cell proliferation by activating p21(WAF/CDKN1A). Biochem Biophys Res Commun. 2020;530:588–96. https://doi.org/10.1016/j.bbrc.2020.07.100.
Lee HJ, Venkatarame Gowda Saralamma V, Kim SM, Ha SE, Vetrivel P, Kim EH, et al. Comparative proteomic profiling of tumor-associated proteins in human gastric cancer cells treated with pectolinarigenin. Nutrients. 2018. https://doi.org/10.3390/nu10111596.
Noyes C, Kitajima S, Li F, Suita Y, Miriyala S, Isaac S, et al. The germline factor DDX4 contributes to the chemoresistance of small cell lung cancer cells. bioRxiv. 2022. https://doi.org/10.1101/2022.04.22.489111.
Ji Xu, Lijie G, Zhenyuan Q, Guangyuan S, Jinming L. ERBB4 promotes the proliferation of gastric cancer cells via the PI3K/Akt signaling pathway. Oncol Rep. 2018;39:2892–8. https://doi.org/10.3892/or.2018.6343.
Huan W, Jianfang R, Qiaoyun Z, Conghua S, Rulin Z, Sihai C, et al. Identification and validation of immune cells and hub genes in gastric cancer microenvironment. Dis Markers. 2022;2022:8639323–8639323. https://doi.org/10.1155/2022/8639323.
Shanshan L, Rujing L, Xiwen L, Daimou Li, Yuzhou Q. Identification and verification of the molecular mechanisms and prognostic values of the cadherin gene family in gastric cancer. Sci Rep. 2021;11:23674–23674. https://doi.org/10.1038/s41598-021-03086-1.
Sanchez-Sandoval AL, Gomora JC. Contribution of voltage-gated sodium channel β-subunits to cervical cancer cells metastatic behavior. Cancer Cell Int. 2019;19:35. https://doi.org/10.1186/s12935-019-0757-6.
Xia J, Huang N, Huang H, Sun L, Dong S, Su J, et al. Voltage-gated sodium channel Nav 1.7 promotes gastric cancer progression through MACC1-mediated upregulation of NHE1. Int J Cancer. 2016;139:2553–69. https://doi.org/10.1002/ijc.30381.
Das K, Gunasegaran B, Tan Iain B, Deng N, Lim KH, Tan P. Mutually exclusive FGFR2, HER2, and KRAS gene amplifications in gastric cancer revealed by multicolour FISH. Cancer Lett. 2014;353:167–75. https://doi.org/10.1016/j.canlet.2014.07.021.
Erdogan S, Aslantas O, Celik S, Atik E. The effects of increased cAMP content on inflammation, oxidative stress and PDE4 transcripts during Brucella melitensis infection. Res Vet Sci. 2008;84:18–25. https://doi.org/10.1016/j.rvsc.2007.02.003.
Ge YJ, Liao QW, Xu YC, Zhao Q, Wu BL, Ye RD. Anti-inflammatory signaling through G protein-coupled receptors. Acta Pharmacol Sin. 2020;41:1531–8. https://doi.org/10.1038/s41401-020-00523-1.
Šedý JR, Ramezani-Rad P. HVEM network signaling in cancer. Adv Cancer Res. 2019;142:145–86. https://doi.org/10.1016/bs.acr.2019.01.004.
Donatelli SS, Djeu JY. Chapter 9 - Immunological sculpting: natural killer-cell receptors and ligands. In: Cancer immunotherapy (Second Edition). Academic Press: San Diego; 2013. p. 115–127. https://doi.org/10.1016/B978-0-12-394296-8.00009-9
Mir MA. Chapter 5 - Costimulation in lymphomas and cancers. In: Developing costimulatory molecules for immunotherapy of diseases. Academic Press; 2015. p. 185–254. https://doi.org/10.1016/B978-0-12-802585-7.00005-4
Szor DJ, Dias AR, Pereira MA, Ramos MFKP, Zilberstein B, Cecconello I, et al. Prognostic role of neutrophil/lymphocyte ratio in resected gastric cancer: a systematic review and meta-analysis. Clinics. 2018;73: e360.
Ting-ting W, Yong-liang Z, Liu-sheng P, Na C, Weisan C, Yi-pin Lv, et al. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut. 2017;66:1900–11.
Bin Z, Guanghua R, Huafeng W, Meng Z, Jianwei Bi, Liye Ma, et al. The prevalence of Th17 cells in patients with gastric cancer. Biochem Biophys Res Commun. 2008;374:533–7. https://doi.org/10.1016/j.bbrc.2008.07.060.
Tao L, Peng Liusheng Yu, Peiwu ZY, Yun S, Xuhu M, et al. Increased circulating Th22 and Th17 cells are associated with tumor progression and patient survival in human gastric cancer. J Clin Immunol. 2012;32:1332–9. https://doi.org/10.1007/s10875-012-9718-8.
Qun Li, Yves L, Tatiana S, Thomas S. Plasmin triggers cytokine induction in human monocyte-derived macrophages. Arteriosclerosis Thromb Vasc Biol. 2007;27:1383–9.
Tatiana S, Marina J, Angela R, Almut S, Thomas S. Plasmin-induced expression of cytokines and tissue factor in human monocytes involves AP-1 and IKKβ-mediated NF-κB activation. Blood. 2001;97:3941–50. https://doi.org/10.1182/blood.V97.12.3941.
Ladislav B, Tatiana S, Thomas S. The serine protease plasmin triggers expression of MCP-1 and CD40 in human primary monocytes via activation of p38 MAPK and Janus kinase (JAK)/STAT signaling pathways. J Biol Chem. 2002;277:33509–17. https://doi.org/10.1074/jbc.M201941200.
Didiasova M, Wujak L, Wygrecka M, Zakrzewicz D. From plasminogen to plasmin: role of plasminogen receptors in human cancer. Int J Mol Sci. 2014;15:21229–52. https://doi.org/10.3390/ijms151121229.
Miles LA, Krajewski S, Baik N, Parmer RJ, Mueller BM. Plg-R(KT) expression in human breast cancer tissues. Biomolecules. 2022. https://doi.org/10.3390/biom12040503.
Miles Lindsey A, Baik N, Krajewski S, Parmer Robert J, Mueller Barbara M. The novel plasminogen receptor, Plg-RKT, and breast cancer progression. Blood. 2011;118:853. https://doi.org/10.1182/blood.V118.21.853.853.
Jin H, Choi H, Kim ES, Lee HH, Cho H, Moon A. Natural killer cells inhibit breast cancer cell invasion through downregulation of urokinase-type plasminogen activator. Oncol Rep. 2021;45:299–308. https://doi.org/10.3892/or.2020.7840.
Funding
Youth Culture Program of First Affiliated Hospital, Anhui Medical University (2020kj04 to SZ), Research Funding for Doctoral Talents of First Affiliated Hospital, Anhui Medical University (1513 to SZ), and Scientific Research of BSKY from Anhui Medical University (XJ201935 to ML).
Author information
Authors and Affiliations
Contributions
HS, JD, ZC and ML: searched data, analyzed data, and wrote the manuscript. RK, XG, SQ: analyzed data. HS, JD, ZC, MH and WH: collect clinical samples and did experiments. SZ and ML: edited the manuscript. SZ and ML: conception of the project.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All procedures performed in studies involving human gastric cancer tissue samples were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from the patient included in this study.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, H., Duan, J., Chen, Z. et al. A prognostic gene signature for gastric cancer and the immune infiltration-associated mechanism underlying the signature gene, PLG. Clin Transl Oncol 25, 995–1010 (2023). https://doi.org/10.1007/s12094-022-03003-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-03003-6