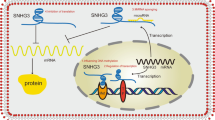
Abstract
Long noncoding RNAs (lncRNA) play pivotal roles in every level of gene and genome regulation. MCM3AP-AS1 is a lncRNA that has an oncogenic role in several kinds of cancers. Aberrant expression of MCM3AP-AS1 has been reported to be involved in the progression of diverse malignancies, including colorectal, cervical, prostate, lymphoma, lung, ovary, liver, bone, and breast cancers. It is generally believed that MCM3AP-AS1 expression is associated with cancer cell growth, proliferation, angiogenesis, and metastasis. MCM3AP-AS1 by targeting various signaling pathways and microRNAs (miRNAs) presents an important role in cancer pathogenesis. MCM3AP-AS1 as a competitive endogenous RNA has the ability to sponge miRNA, inhibit their expressions, and bind to different target mRNAs related to cancer development. Therefore, MCM3AP-AS1 by targeting several signaling pathways, including the FOX family, Wnt, EGF, and VEGF can be a potent target for cancer prediction and diagnosis. In this review, we will summarize the role of MCM3AP-AS1 in various human cancers.

Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AGGF1:
-
Angiogenic factor with G-patch and FHA domain 1
- BL:
-
Burkitt lymphoma
- BC:
-
Breast cancer
- CENPF:
-
Centromere protein F
- CSCC:
-
Squamous cell carcinoma
- CDK:
-
Cyclin-dependent kinase
- CCND1:
-
Cyclin D1
- ccRCC:
-
Clear cell renal cell carcinoma
- ceRNA:
-
Competing endogenous
- CRC:
-
Colorectal cancer
- DDK:
-
Dbf4 dependent kinase
- DPP:
-
Dipeptidyl peptidase
- EIF4E:
-
Eukaryotic translation initiation factor 4E
- ER:
-
Estrogen receptor
- EC:
-
Endometrial cancer
- EMT:
-
Epithelial–mesenchymal transition
- Erα:
-
Estrogen receptor α
- FOXF2:
-
Forkhead box F2
- GBM:
-
Glioblastoma
- GC:
-
Gastric cancer
- HCC:
-
Hepatocellular carcinoma
- HPV:
-
Human papillomavirus
- lncRNA:
-
Long noncoding RNAs
- miRNAs:
-
MicroRNAs
- MCM:
-
Minichromosome maintenance protein
- MMP1:
-
Matrix metalloproteinase 1
- NHL:
-
Non-Hodgkin lymphoma
- ORC:
-
Origin recognition complex
- OSCC:
-
Oral squamous cell carcinoma
- OC:
-
Ovarian cancer
- pre-RC:
-
Pre-replicative complex
- PARP:
-
Poly (ADP-ribose) polymerase 1
- PaC:
-
Pancreatic cancer
- PTC:
-
Papillary thyroid cancer
- PrC:
-
Prostate cancer
- PR:
-
Progesterone receptor
- RCC:
-
Renal cell carcinoma
- SPARC:
-
Secreted protein acidic and cysteine rich
- TAK1:
-
Transforming growth factor‑β‑activated kinase 1
- VEGF-A:
-
Vascular endothelial growth factor-A
- ZFP36:
-
RNA-binding protein ZFP36 ring finger protein
References
Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573.
Sun Y-M, Chen Y-Q. Principles and innovative technologies for decrypting noncoding RNAs: from discovery and functional prediction to clinical application. J Hematol Oncol. 2020;13:109.
Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, Balligand J-L, Devaux Y. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids. 2017;8:494–507.
Wong NK, Huang C-L, Islam R, Yip SP. Long non-coding RNAs in hematological malignancies: translating basic techniques into diagnostic and therapeutic strategies. J Hematol Oncol. 2018;11:131.
Youness RA, Gad MZ. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4:36–44.
Pecero ML, Salvador-Bofill J, Molina-Pinelo S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol. 2019;42:1–12.
Dieter C, Lemos NE, Corrêa NRdF, Assmann TS, Crispim D. The impact of lncRNAs in diabetes mellitus: a systematic review and in silico analyses. Front Endocrinol. 2021;12:602597.
Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592:2884–900.
Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X, Cui Y. Long non-coding RNA MIAT in development and disease: a new player in an old game. J Biomed Sci. 2018;25:23.
Karakas D, Ozpolat B. The role of LncRNAs in translation. Noncoding RNA. 2021;7:16.
Yu Y, Lai S, Peng X. Long non-coding RNA MCM3AP-AS1 facilitates colorectal cancer progression by regulating the microRNA-599/ARPP19 axis. Oncol Lett. 2021;21:1–1.
Lan L, Liang Z, Zhao Y, Mo Y: 2020 LncRNA MCM3AP-AS1 inhibits cell proliferation in cervical squamous cell carcinoma by down-regulating miRNA-93. Bioscience reports 40.
Bharti D, Jang S-J, Lee S-Y, Lee S-L, Rho G-J. In vitro generation of oocyte like cells and their in vivo efficacy: how far we have been succeeded. Cells. 2020;9:557.
Sun H, Wu P, Zhang B, Wu X, Chen W. MCM3AP-AS1 promotes cisplatin resistance in gastric cancer cells via the miR-138/FOXC1 axis. Oncol Lett. 2021;21:1–1.
Chen Y, Chen Z, Mo J, Pang M, Chen Z, Feng F, Xie P, Yang B. Identification of HCG18 and MCM3AP-AS1 that associate with bone metastasis, poor prognosis and increased abundance of M2 macrophage infiltration in prostate cancer. Technol Cancer Res Treat. 2021;20:1533033821990064.
Guo C, Gong M, Li Z. Knockdown of lncRNA MCM3AP-AS1 attenuates chemoresistance of burkitt lymphoma to doxorubicin treatment via targeting the miR-15a/EIF4E axis. Cancer Manag Res. 2020;12:5845.
Yang C, Zheng J, Xue Y, Yu H, Liu X, Ma J, Liu L, Wang P, Li Z, Cai H. The effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front Mol Neurosci. 2018;10:437.
Li X, Yu M, Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J Cell Biochem. 2020;121:2258–67.
Wen J, Han S, Cui M, Wang Y. Long non-coding RNA MCM3AP-AS1 drives ovarian cancer progression via the microRNA-143-3p/TAK1 axis. Oncol Rep. 2020;44:1375–84.
Zhang H, Luo C, Zhang G. LncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting with miR-455. DNA Cell Biol. 2019;38:857–64.
Zhang H, Lu B. The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers. 2020;12:2926.
Chen Q, Xu H, Zhu J, Feng K, Hu C. LncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed Pharmacother. 2020;128: 110289.
Yan Y, Yu J, Liu H, Guo S, Zhang Y, Ye Y, Xu L, Ming L. Construction of a long non-coding RNA-associated ceRNA network reveals potential prognostic lncRNA biomarkers in hepatocellular carcinoma. Pathol Res Pract. 2018;214:2031–8.
Ying H, Ebrahimi M, Keivan M, Khoshnam SE, Salahi S, Farzaneh M. miRNAs; a novel strategy for the treatment of COVID-19. Cell Biol Int. 2021;45:2045–53.
Anbiyaiee A, Ramazii M, Bajestani SS, Meybodi SM, Keivan M, Khoshnam SE, Farzaneh M: 2022 The function of LncRNA-ATB in cancer. Clin Transl Oncol 1–9.
Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509.
Shah IM, Dar MA, Bhat KA, Dar TA, Ahmad F, Ahmad SM: Long Non-Coding RNAs: Biogenesis, Mechanism of Action and Role in Different Biological and Pathological Processes. 2022.
Soudyab M, Iranpour M, Ghafouri-Fard S: The role of long non-coding RNAs in breast cancer. Archives of Iranian medicine 2016, 19:0–0.
An C, Wang I, Li X, Xia R, Deng F. Long non-coding RNA in prostate cancer. Am J Clin Exp Urol. 2022;10:170–9.
Xu W-W, Jin J, Wu X-Y, Ren Q-L, Farzaneh M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 2022;22:1–9.
Li L, Bi Y, Diao S, Li X, Yuan T, Xu T, Huang C, Li J. Exosomal LncRNAs and hepatocellular carcinoma: from basic research to clinical practice. Biochem Pharmacol. 2022;200:115032.
Wang W, Xiang M, Liu H, Chu X, Sun Z, Feng L. A prognostic risk model based on DNA methylation levels of genes and lncRNAs in lung squamous cell carcinoma. PeerJ. 2022;10: e13057.
Li X, Lv J, Liu S. MCM3AP-AS1 KD inhibits proliferation, invasion, and migration of PCa cells via DNMT1/DNMT3 (A/B) methylation-mediated upregulation of NPY1R. Mol Ther Nucleic Acids. 2020;20:265–78.
Yu X, Zheng Q, Zhang Q, Zhang S, He Y, Guo W. MCM3AP-AS1: an indispensable cancer-related LncRNA. Front Cell Dev Biol. 2021;9: 752718.
Chowdhary A, Satagopam V, Schneider R. Long non-coding RNAs: mechanisms, experimental, and computational approaches in identification, characterization, and their biomarker potential in cancer. Front Genet. 2021;12:770.
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:1–16.
Jia Z, Li W, Bian P, Liu H, Pan D, Dou Z. LncRNA MCM3AP-AS1 promotes cell proliferation and invasion through regulating miR-543-3p/SLC39A10/PTEN axis in prostate cancer. Onco Targets Ther. 2020;13:9365.
Shen D, Li J, Tao K, Jiang Y. Long non-coding RNA MCM3AP antisense RNA 1 promotes non-small cell lung cancer progression through targeting microRNA-195-5p. Bioengineered. 2021;12:3525–38.
Ma X, Luo J, Zhang Y, Sun D, Lin Y. LncRNA MCM3AP-AS1 upregulates CDK4 by sponging miR-545 to suppress G1 Arrest in colorectal cancer. Cancer Manag Res. 2020;12:8117.
Jiang L, Hu W, Yao N: Prognostic Impact of lncRNA MCM3AP-AS1 Expression in Malignant Solid Tumors: A Systematic Review and Meta-analysis. 2021.
Wu J, Lv Y, Li Y, Jiang Y, Wang L, Zhang X, Sun M, Zou Y, Xu J, Zhang L. MCM3AP-AS1/miR-876-5p/WNT5A axis regulates the proliferation of prostate cancer cells. Cancer Cell Int. 2020;20:1–12.
Yang M, Sun S, Guo Y, Qin J, Liu G. Long non-coding RNA MCM3AP-AS1 promotes growth and migration through modulating FOXK1 by sponging miR-138-5p in pancreatic cancer. Mol Med. 2019;25:1–10.
Crombie J, LaCasce A. The treatment of Burkitt lymphoma in adults. Blood J Am Soc Hematol. 2021;137:743–50.
Casulo C, Friedberg JW. Burkitt lymphoma-a rare but challenging lymphoma. Best Pract Res Clin Haematol. 2018;31:279–84.
Kalisz K, Alessandrino F, Beck R, Smith D, Kikano E, Ramaiya NH, Tirumani SH. An update on Burkitt lymphoma: a review of pathogenesis and multimodality imaging assessment of disease presentation, treatment response, and recurrence. Insights Imaging. 2019;10:1–16.
Wang Q-M, Lian G-Y, Song Y, Huang Y-F, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019;231:116335.
Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z, Yeoh AE, Tenen DG, Sanda T. Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia. 2018;32:2138–51.
Huang P-S, Chung I, Lin Y-H, Lin T-K, Chen W-J, Lin K-H. The long non-coding RNA MIR503HG enhances proliferation of human ALK-negative anaplastic large-cell lymphoma. Int J Mol Sci. 2018;19:1463.
Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16:1–13.
Tarantelli C, Lupia A, Stathis A, Bertoni F. Is there a role for dual PI3K/mTOR inhibitors for patients affected with lymphoma? Int J Mol Sci. 2020;21:1060.
Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans. 2015;43:763–72.
Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:1–19.
Bitterman PB, Polunovsky VA. eIF4E-mediated translational control of cancer incidence. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2015;1849:774–80.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:1–19.
Blondeau JJ, Deng M, Syring I, Schrödter S, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics. 2015;7:1–10.
Gao Y, Li H, Ma X, Fan Y, Ni D, Zhang Y, Huang Q, Liu K, Li X, Wang L. KLF6 suppresses metastasis of clear cell renal cell carcinoma via transcriptional repression of E2F1. Can Res. 2017;77:330–42.
Enz N, Vliegen G, De Meester I, Jungraithmayr W. CD26/DPP4-a potential biomarker and target for cancer therapy. Pharmacol Ther. 2019;198:135–59.
Larrinaga G, Blanco L, Sanz B, Perez I, Gil J, Unda M, Andrés L, Casis L, López JI. The impact of peptidase activity on clear cell renal cell carcinoma survival. Am J Physiol Renal Physiol. 2012;303:F1584–91.
Qiu L, Ma Y, Yang Y, Ren X, Wang D, Jia X. Pro-angiogenic and pro-inflammatory regulation by lncRNA MCM3AP-AS1-mediated upregulation of DPP4 in clear cell renal cell carcinoma. Front Oncol. 2020;10:705.
Shan S, Wang Y, Zhu C. A comprehensive expression profile of tRNA-derived fragments in papillary thyroid cancer. J Clin Lab Anal. 2021;35:e23664.
Peng X, Zhang K, Ma L, Xu J, Chang W. The role of long non-coding RNAs in thyroid cancer. Front Oncol. 2020;10:941–941.
Liang M, Jia J, Chen L, Wei B, Guan Q, Ding Z, Yu J, Pang R, He G. LncRNA MCM3AP-AS1 promotes proliferation and invasion through regulating miR-211-5p/SPARC axis in papillary thyroid cancer. Endocrine. 2019;65:318–26.
Tsui DCC, Camidge DR, Rusthoven CG. Managing central nervous system spread of lung cancer: the state of the art. J Clin Oncol. 2022;40:642–60.
Chen Y, Zitello E, Guo R, Deng Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin Transl Med. 2021;11:e367–e367.
Jiang J, Lu Y, Zhang F, Huang J, Ren X-l, Zhang R: The Emerging Roles of Long Noncoding RNAs as Hallmarks of Lung Cancer. Frontiers in Oncology 2021, 11.
Lin N-C, Hsien S-I, Hsu J-T, Chen MYC. Impact on patients with oral squamous cell carcinoma in different anatomical subsites: a single-center study in Taiwan. Sci Rep. 2021;11:15446.
Hema K, Smitha T, Sheethal H, Mirnalini SA. Epigenetics in oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2017;21:252.
Mourad M, Jetmore T, Jategaonkar AA, Moubayed S, Moshier E, Urken ML. Epidemiological trends of head and neck cancer in the United States: a SEER population study. J Oral Maxillofac Surg. 2017;75:2562–72.
Nakashima C, Yamamoto K, Fujiwara-Tani R, Luo Y, Matsushima S, Fujii K, Ohmori H, Sasahira T, Sasaki T, Kitadai Y. Expression of cytosolic malic enzyme (ME 1) is associated with disease progression in human oral squamous cell carcinoma. Cancer Sci. 2018;109:2036–45.
Zhuang Z, Yu P, Xie N, Wu Y, Liu H, Zhang M, Tao Y, Wang W, Yin H, Zou B. MicroRNA-204-5p is a tumor suppressor and potential therapeutic target in head and neck squamous cell carcinoma. Theranostics. 2020;10:1433.
Kong X-P, Yao J, Luo W, Feng F-K, Ma J-T, Ren Y-P, Wang D-l, Bu R-F. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol Cell Biochem. 2014;394:177–86.
Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–7.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Przeglad Gastroenterologiczny. 2019;14:89–103.
Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17.
Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu J, Yue T, Wang J, Zhu J, Bu D. The novel long noncoding RNA CRART16 confers cetuximab resistance in colorectal cancer cells by enhancing ERBB3 expression via miR-371a-5p. Cancer Cell Int. 2020;20:1–16.
Yang Z, Zhang J, Lu D, Sun Y, Zhao X, Wang X, Zhou W, He Q, Jiang Z. Hsa_circ_0137008 suppresses the malignant phenotype in colorectal cancer by acting as a microRNA-338-5p sponge. Cancer Cell Int. 2020;20:1–12.
Zhang J, Bian Z, Jin G, Liu Y, Li M, Yao S, Zhao J, Feng Y, Wang X, Yin Y. Long non-coding RNA IQCJ-SCHIP1 antisense RNA 1 is downregulated in colorectal cancer and inhibits cell proliferation. Ann Transl Med. 2019;7:198.
Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, Yao S, Jin G, Du J, Han W. LncRNA–FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24:4808–19.
Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6:1–11.
Gulimiheranmu M, Wang X, Zhou J. Advances in female germ cell induction from pluripotent stem cells. Stem Cells Int. 2021;2021:1–13.
Zhang P, Ji D-B, Han H-B, Shi Y-F, Du C-Z, Gu J. Downregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2014;20:12241.
Chen MC, Nhan DC, Hsu CH, Wang TF, Li CC, Ho TJ, Mahalakshmi B, Chen MC, Yang LY, Huang CY. SENP1 participates in Irinotecan resistance in human colon cancer cells. J Cell Biochem. 2021;122:1277–94.
Dai W, Zeng W, Lee D. lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. J Gene Med. 2021;23: e3306.
Yu F-B, Sheng J, Yu J-M, Liu J-H, Qin X-X, Mou B. MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J Gastroenterol. 2020;26:627.
Zhang J, Zhang C, Sang L, Huang L, Du J, Zhao X. FOXF2 inhibits proliferation, migration, and invasion of Hela cells by regulating Wnt signaling pathway. Biosci Rep. 2018;38:BSR20180747.
Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678–88.
Gu J-F, Wang G-Q, Gao Y-C, Dai Y-J. Overexpression of miR-599 is associated with metastasis in colorectal cancer via inhibition of SATB2. Int J Clin Exp Pathol. 2017;10:6701–9.
Song H, Pan J, Liu Y, Wen H, Wang L, Cui J, Liu Y, Hu B, Yao Z, Ji G. Increased ARPP-19 expression is associated with hepatocellular carcinoma. Int J Mol Sci. 2015;16:178–92.
Horiuchi A, Williams K, Kurihara T, Nairn A, Greengard P. Purification and cDNA cloning of ARPP-16, a cAMP-regulated phosphoprotein enriched in basal ganglia, and of a related phosphoprotein, ARPP-19. J Biol Chem. 1990;265:9476–84.
Du B, Wang Z, Zhang X, Feng S, Wang G, He J, Zhang B. MicroRNA-545 suppresses cell proliferation by targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One. 2014;9:e88022.
Wu A, Wu B, Guo J, Luo W, Wu D, Yang H, Zhen Y, Yu X, Wang H, Zhou Y. Elevated expression of CDK4 in lung cancer. J Transl Med. 2011;9:1–9.
Ching Y-P, Leong VY, Lee M-F, Xu H-T, Jin D-Y. Ng IO-L: P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation. Can Res. 2007;67:3601–8.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604.
Sun Z, Zhao L, Wang S, Wang H. Knockdown of long non-coding RNA LINC01006 represses the development of hepatocellular carcinoma by modulating the miR-194-5p/CADM1 axis. Ann Hepatol. 2021;27:100571.
Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu Y, Han J, Zhang M. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495:1074–80.
Wang B, Zou A, Ma L, Chen X, Wang L, Zeng X, Tan T. miR-455 inhibits breast cancer cell proliferation through targeting CDK14. Eur J Pharmacol. 2017;807:138–43.
Zhang L, Li X, Lu J, Qian Y, Qian T, Wu X, Xu Q. The EGFR polymorphism increased the risk of hepatocellular carcinoma through the miR-3196-dependent approach in Chinese Han population. Pharmacogenomics Pers Med. 2021;14:469.
Ning T, Peng Z, Li S, Qu Y, Zhang H, Duan J, Wang X, Yang H, Liu R, Deng T. miR-455 inhibits cell proliferation and migration via negative regulation of EGFR in human gastric cancer. Oncol Rep. 2017;38:175–82.
Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang X-S, Bannon F, Ahn JV, Johnson CJ, Bonaventure A. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2). The Lancet. 2015;385:977–1010.
Alberts S, Cervantes A, Van de Velde C. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol. 2003;14:31–6.
Ashraf N, Hoffe S, Kim R. Adjuvant treatment for gastric cancer: chemotherapy versus radiation. Oncologist. 2013;18:1013.
Li C-Y, Liang G-Y, Yao W-Z, Sui J, Shen X, Zhang Y-Q, Peng H, Hong W-W, Ye Y-C, Zhang Z-Y. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48:1965–76.
Monteleone NJ, Lutz CS. miR-708-5p: a microRNA with emerging roles in cancer. Oncotarget. 2017;8:71292.
Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan IB, Gopalakrishnan V, Ooi CH, Lee J, Qin L, Wu J. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut. 2013;62:22–33.
Dai Q, Zhang T, Pan J, Li C. LncRNA UCA1 promotes cisplatin resistance in gastric cancer via recruiting EZH2 and activating PI3K/AKT pathway. J Cancer. 2020;11:3882.
Yang Z, Jiang S, Cheng Y, Li T, Hu W, Ma Z, Chen F, Yang Y. FOXC1 in cancer development and therapy: deciphering its emerging and divergent roles. Ther Adv Med Oncol. 2017;9:797–816.
Barbato S, Solaini G, Fabbri M. MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol. 2017;333:229–68.
Glehen O, Gilly FN, Arvieux C, Cotte E, Boutitie F, Mansvelt B, Bereder JM, Lorimier G, Quenet F, Elias D. Peritoneal carcinomatosis from gastric cancer: a multi-institutional study of 159 patients treated by cytoreductive surgery combined with perioperative intraperitoneal chemotherapy. Ann Surg Oncol. 2010;17:2370–7.
Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J. LncRNAs in pancreatic cancer. Oncotarget. 2016;7:57379.
Zhao Y-X, Chen S-R, Su P-P, Huang F-H, Shi Y-C, Shi Q-Y, Lin S. Using mesenchymal stem cells to treat female infertility: an update on female reproductive diseases. Stem Cells Int. 2019;2019:9071720–9071720.
Wencong M, Jinghan W, Yong Y, Jianyang A, Bin L, Qingbao C, Chen L, Xiaoqing J. FOXK1 promotes proliferation and metastasis of gallbladder cancer by activating AKT/mTOR signaling pathway. Front Oncol. 2020;10:545.
Yang M, Sun S, Guo Y, Qin J, Liu G. Long non-coding RNA MCM3AP-AS1 promotes growth and migration through modulating FOXK1 by sponging miR-138-5p in pancreatic cancer. Mol Med. 2019;25:55.
Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18:3–9.
Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs. 2016;20:S2–8.
Zeng T, Li L, Zhou Y, Gao L. Exploring long noncoding RNAs in glioblastoma: regulatory mechanisms and clinical potentials. Int J Genomics. 2018;2018:2895958–2895958.
Yadav B, Pal S, Rubstov Y, Goel A, Garg M, Pavlyukov M, Pandey AK. LncRNAs associated with glioblastoma: from transcriptional noise to novel regulators with a promising role in therapeutics. Mol Ther Nucleic Acids. 2021;24:728–42.
Ji Y, Gu Y, Hong S, Yu B, Zhang JH, Liu JN. Comprehensive analysis of lncRNA-TF crosstalks and identification of prognostic regulatory feedback loops of glioblastoma using lncRNA/TF-mediated ceRNA network. J Cell Biochem. 2020;121:755–67.
Rawla P. Epidemiology of prostate cancer. World J Oncol. 2019;10:63–89.
Jiang G, Su Z, Liang X, Huang Y, Lan Z, Jiang X. Long non-coding RNAs in prostate tumorigenesis and therapy. Mol Clin Oncol. 2020;13:1–1.
Wong SK, Mohamad N-V, Giaze TR, Chin K-Y, Mohamed N, Ima-Nirwana S. Prostate cancer and bone metastases: the underlying mechanisms. Int J Mol Sci. 2019;20:2587.
Wu J, Lv Y, Li Y, Jiang Y, Wang L, Zhang X, Sun M, Zou Y, Xu J, Zhang L. MCM3AP-AS1/miR-876-5p/WNT5A axis regulates the proliferation of prostate cancer cells. Cancer Cell Int. 2020;20:307.
Ren Z, Yang T, Zhang P, Liu K, Liu W, Wang P. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep. 2019;19:515–23.
Yu C, Cao H, He X, Sun P, Feng Y, Chen L, Gong H. Cyclin-dependent kinase inhibitor 3 (CDKN3) plays a critical role in prostate cancer via regulating cell cycle and DNA replication signaling. Biomed Pharmacother. 2017;96:1109–18.
Yaqinuddin A, Qureshi SA, Qazi R, Farooq S, Abbas F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J Urol. 2009;182:756–61.
Cavaliere AF, Perelli F, Zaami S, Piergentili R, Mattei A, Vizzielli G, Scambia G, Straface G, Restaino S, Signore F: Towards Personalized Medicine: Non-Coding RNAs and Endometrial Cancer. In Healthcare. Multidisciplinary Digital Publishing Institute; 2021: 965.
Piura E, Piura B. Brain metastases from endometrial carcinoma. Int Sch Res Notices. 2012;2012:1–13.
Rosa-Rosa JM, Leskelä S, Cristóbal-Lana E, Santón A, López-García MÁ, Muñoz G, Pérez-Mies B, Biscuola M, Prat J, Esther OE. Molecular genetic heterogeneity in undifferentiated endometrial carcinomas. Mod Pathol. 2016;29:1390–8.
Yu J, Fan Q, Li L. The MCM3AP-AS1/miR-126/VEGF axis regulates cancer cell invasion and migration in endometrioid carcinoma. World J Surg Oncol. 2021;19:1–8.
Zhang X, Zeng Q, Cai W, Ruan W. Trends of cervical cancer at global, regional, and national level: data from the Global Burden of Disease study 2019. BMC Public Health. 2021;21:894.
Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–99.
Small W Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR, Viswanathan AN. Cervical cancer: a global health crisis. Cancer. 2017;123:2404–12.
Aggarwal P. Cervical cancer: can it be prevented? World J Clin Oncol. 2014;5:775–80.
Zhang Y, Luo G, Li M, Guo P, Xiao Y, Ji H, Hao Y. Global patterns and trends in ovarian cancer incidence: age, period and birth cohort analysis. BMC Cancer. 2019;19:984.
Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:1–22.
Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res. 2018;37:1–13.
Łukasiewicz S, Czeczelewski M, Forma A, Baj J, Sitarz R, Stanisławek A. Breast cancer—epidemiology, risk factors, classification, prognostic markers, and current treatment strategies—an updated review. Cancers. 2021;13:4287.
Simone V, D’Avenia M, Argentiero A, Felici C, Rizzo FM, De Pergola G, Silvestris F. Obesity and breast cancer: molecular interconnections and potential clinical applications. Oncologist. 2016;21:404–17.
Loftus KM, Cui H, Coutavas E, King DS, Ceravolo A, Pereiras D, Solmaz SR. Mechanism for G2 phase-specific nuclear export of the kinetochore protein CENP-F. Cell Cycle. 2017;16:1414–29.
Tang T-P, Qin C-X, Yu H. MCM3AP-AS1 regulates proliferation, apoptosis, migration, and invasion of breast cancer cells via binding with ZFP36. Trans Cancer Res. 2021;10:4478–88.
Sun P, Feng Y, Guo H, Li R, Yu P, Zhou X, Pan Z, Liang Y, Yu B, Zheng Y. MiR-34a inhibits cell proliferation and induces apoptosis in human nasopharyngeal carcinoma by targeting lncRNA MCM3AP-AS1. Cancer Manag Res. 2020;12:4799.
Zhou M, Bian Z, Liu B, Zhang Y, Cao Y, Cui K, Sun S, Li J, Zhang J, Wang X. Long noncoding RNA MCM3AP-AS1 enhances cell proliferation and metastasis in colorectal cancer by regulating miR-193a-5p/SENP1. Cancer Med. 2021;10:2470–81.
Riahi A, Hosseinpour-Feizi M, Rajabi A, Akbarzadeh M, Montazeri V, Safaralizadeh R. Overexpression of long non-coding RNA MCM3AP-AS1 in breast cancer tissues compared to adjacent non-tumour tissues. Br J Biomed Sci. 2021;78:53–7.
Tchounwou PB, Dasari S, Noubissi FK, Ray P, Kumar S. Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. J Exp Pharmacol. 2021;13:303.
Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–83.
Bierhoff H: Analysis of lncRNA-protein interactions by RNA-protein pull-down assays and RNA immunoprecipitation (RIP). In Cellular Quiescence. Springer; 2018: 241–250
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein–lncRNA interaction. Brief Bioinform. 2016;17:106–16.
Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291.
Wang Y, Zhang M, Zhou F. Biological functions and clinical applications of exosomal long non-coding RNAs in cancer. J Cell Mol Med. 2020;24:11656–66.
Zhao W, Liu Y, Zhang C, Duan C. Multiple roles of exosomal long noncoding RNAs in cancers. Biomed Res Int. 2019;2019:1460572.
Lang H, Hu G, Chen Y, Liu Y, Tu W, Lu Y, Wu L, Xu G. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci. 2017;21:959–72.
Dong P, Xiong Y, Yue J, Hanley JBS, Kobayashi N, Todo Y, Watari H. Exploring lncRNA-mediated regulatory networks in endometrial cancer cells and the tumor microenvironment: advances and challenges. Cancers. 2019;11:234.
Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-targeted therapeutics. Cell Metab. 2018;27:714–39.
Roberts TC, Langer R, Wood MJ. Advances in oligonucleotide drug delivery. Nat Rev Drug Discovery. 2020;19:673–94.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom Proteom Bioinf. 2016;14:42–54.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9.
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69.
Jiang M-C, Ni J-J, Cui W-Y, Wang B-Y, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354.
Author information
Authors and Affiliations
Contributions
SHA, FGH, MSH, SHU, MGH, AM, MK, MCH, and ZN have made contributions to the writing of the manuscript. SE. KH., and M. F. have made contributions to the design of the figure and the revision of the manuscript. All authors have approved the submitted version of the article and have agreed to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Azizidoost, S., Ghaedrahmati, F., Sheykhi-Sabzehpoush, M. et al. The role of LncRNA MCM3AP-AS1 in human cancer. Clin Transl Oncol 25, 33–47 (2023). https://doi.org/10.1007/s12094-022-02904-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02904-w