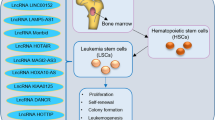
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is a malignancy caused by clonal proliferation of T-cell pre-cursors arising from the thymus. Although the optimized chemotherapy regimen could improve the outcome of such patients, some challenges such as higher risk for induction failure, early relapse and isolated central nervous system (CNS) relapse occurring in T-ALL patients are of great significance, leading to increased mortality rates. Long non-coding RNA (lncRNA) is a key component involved in cell signaling through a variety of mechanisms in regulating gene expression. Oncogenes and tumor suppressors are no exception and their expression can be affected by lncRNAs. In addition, accumulating researches in samples from T-ALL patients as well as pre-clinical studies in mice suggest that the expression profile of lncRNAs in T-ALL could be aberrant, resulting in deregulation of target genes and downstream signaling pathways. In addition, accumulating researches in samples from T-ALL patients as well as pre-clinical studies in mice suggest that the expression profile of lncRNAs in T-ALL could be aberrant, resulting in deregulation of target genes and downstream signaling pathways. These lncRNAs may be determinants of proliferation, apoptosis, and drug resistance observed in T-ALL. Thus, lncRNAs can be a good tool to develop novel strategies against cancer cells in the treatment of relapsed and refractory T-ALL. They can also act as promoting biomarkers in assessing T-ALL and differentiating between patients with poor prognosis and good prognosis.

Similar content being viewed by others
References
Pulsipher MA, Langholz B, Wall DA, Schultz KR, Bunin N, Carroll W, et al. Risk factors and timing of relapse after allogeneic transplantation in pediatric ALL: for whom and when should interventions be tested? Bone Marrow Transplant. 2015;50(9):1173–9.
Ekiz HA, Can G, Baran Y. Role of autophagy in the progression and suppression of leukemias. Crit Rev Oncol Hematol. 2012;81(3):275–85.
Kahn JM, Keegan TH, Tao L, Abrahão R, Bleyer A, Viny AD. Racial disparities in the survival of American children, adolescents, and young adults with acute lymphoblastic leukemia, acute myelogenous leukemia, and Hodgkin lymphoma. Cancer. 2016;122(17):2723–30.
Yang J, Yang Y. Long noncoding RNA endogenous bornavirus-like nucleoprotein acts as an oncogene by regulating microRNA-655-3p expression in T-cell acute lymphoblastic leukemia. Bioengineered. 2022;13(3):6409–19.
Liu Q, Ma H, Sun X, Liu B, Xiao Y, Pan S, et al. The regulatory ZFAS1/miR-150/ST6GAL1 crosstalk modulates sialylation of EGFR via PI3K/Akt pathway in T-cell acute lymphoblastic leukemia. J Exp Clin Cancer Res. 2019;38(1):1–15.
Nashwa E-K, Aziz MAA, Hesham M, Matbouly S, Mostafa SA, Bakkar A, et al. Upregulation of leukemia-induced non-coding activator RNA (LUNAR1) predicts poor outcome in pediatric T-acute lymphoblastic leukemia. Immunobiology. 2021;226(6): 152149.
Mahmoudian-Sani M-R, Jalali A, Jamshidi M, Moridi H, Alghasi A, Shojaeian A, et al. Long non-coding RNAs in thyroid cancer: implications for pathogenesis, diagnosis, and therapy. Oncol Res Treatm. 2019;42(3):136–42.
Carninci P, Kasukawa T, Katayama S, Gough J, Frith M, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63.
Mattick JS. Editor Long noncoding RNAs in cell and developmental biology. Seminars Cell Develop Biol. 2011;22:327.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–19.
Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev. 2013;33(3):517–53.
Kumar M, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Topic Med Chem. 2017;17(15):1750–7.
Wang Y, Wu P, Lin R, Rong L, Xue Y, Fang Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci Rep. 2015;5(1):1–10.
Kermezli Y, Saadi W, Belhocine M, Mathieu E-L, Garibal M-A, Asnafi V, et al. A comprehensive catalog of LncRNAs expressed in T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2019;60(8):2002–14.
Sartori DA, Chan DW. Biomarkers in prostate cancer: what’s new? Curr Opin Oncol. 2014;26(3):259.
Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113(1):1–11.
Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7(3):761–6.
Da Sacco L, Baldassarre A, Masotti A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci. 2011;13(1):97–114.
Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood J Am Soc Hematol. 2007;109(12):5191–8.
Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–9.
Wallaert A, Durinck K, Taghon T, Van Vlierberghe P, Speleman F. T-ALL and thymocytes: a message of noncoding RNAs. J Hematol Oncol. 2017;10(1):1–17.
Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107(6):2223–33.
González-García S, García-Peydró M, Martín-Gayo E, Ballestar E, Esteller M, Bornstein R, et al. CSL–MAML-dependent Notch1 signaling controls T lineage-specific IL-7Rα gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206(4):779–91.
Weng AP, Ferrando AA, Lee W, Morris JP IV, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71.
Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606.
Peng W, Feng J. Long noncoding RNA LUNAR1 associates with cell proliferation and predicts a poor prognosis in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;77:65–71.
Kelliher MA, Roderick JE. NOTCH signaling in T-cell-mediated anti-tumor immunity and T-cell-based immunotherapies. Front Immunol. 2018;9:1718.
Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood J AmSoc Hematol. 2017;129(9):1113–23.
Gusscott S, Tamiro F, Giambra V, Weng AP. Insulin-like growth factor (IGF) signaling in T-cell acute lymphoblastic leukemia. Adv Biol Regul. 2019;74: 100652.
Medyouf H, Gusscott S, Wang H, Tseng J-C, Wai C, Nemirovsky O, et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med. 2011;208(9):1809–22.
Piao H-Y, Guo S, Wang Y, Zhang J. Long noncoding RNA NALT1-induced gastric cancer invasion and metastasis via NOTCH signaling pathway. World J Gastroenterol. 2019;25(44):6508.
Li Y, Rui X, Chen D, Xuan H, Yang H, Meng X. LncRNA AWPPH as a prognostic predictor in human cancers: evidence from meta-analysis. Oncol Lett. 2020. https://doi.org/10.3892/ol.2020.12102.
Li X, Song F, Sun H. Long non-coding RNA AWPPH interacts with ROCK2 and regulates the proliferation and apoptosis of cancer cells in pediatric T-cell acute lymphoblastic leukemia. Oncol Lett. 2020;20(5):1.
Nakagawa O, Fujisawa K, Ishizaki T, Saito Y, Nakao K, Narumiya S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996;392(2):189–93.
Wang L, Hou G, Xue L, Li J, Wei P, Xu P. Autocrine motility factor receptor signaling pathway promotes cell invasion via activation of ROCK-2 in esophageal squamous cell cancer cells. Cancer Invest. 2010;28(10):993–1003.
Gao C, Zhang J, Wang Q, Ren C. Overexpression of lncRNA NEAT1 mitigates multidrug resistance by inhibiting ABCG2 in leukemia. Oncol Lett. 2016;12(2):1051–7.
Huang X, Zhong R, He X, Deng Q, Peng X, Li J, et al. Investigations on the mechanism of progesterone in inhibiting endometrial cancer cell cycle and viability via regulation of long noncoding RNA NEAT1/microRNA-146b-5p mediated Wnt/β-catenin signaling. IUBMB Life. 2019;71(2):223–34.
Correia NC, Fragoso R, Carvalho T, Enguita FJ, Barata JT. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci Rep. 2016;6(1):1–10.
Luo Y-Y, Wang Z-H, Yu Q, Yuan L-L, Peng H-L, Xu Y-X. LncRNA-NEAT1 promotes proliferation of T-ALL cells via miR-146b-5p/NOTCH1 signaling pathway. Pathol Res Pract. 2020;216(11): 153212.
Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z, Yeoh A, et al. Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia. 2018;32(10):2138–51.
Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 2011;30(3):494–509.
Lécuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004;32(1):11–24.
Leong WZ, Tan SH, Ngoc PCT, Amanda S, Yam AWY, Liau W-S, et al. ARID5B as a critical downstream target of the TAL1 complex that activates the oncogenic transcriptional program and promotes T-cell leukemogenesis. Genes Dev. 2017;31(23–24):2343–60.
Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MC, et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood J Am Soc Hematol. 2019;134(3):239–51.
Salvesen GS, Riedl SJ. Caspase mechanisms. Programmed cell death in cancer progression and therapy. 2008:13–23.
Chaudhry P, Singh M, Parent S, Asselin E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol Cell Biol. 2012;32(4):826–39.
Zhang L, Xu H-G, Lu C. A novel long non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to induce cellular apoptosis in T-cell acute lymphoblastic leukemia cells. Leuk Lymphoma. 2014;55(6):1373–82.
Candé C, Cohen I, Daugas E, Ravagnan L, Larochette N, Zamzami N, et al. Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie. 2002;84(2–3):215–22.
Fan F-Y, Deng R, Yi H, Sun H-P, Zeng Y, He G-C, et al. The inhibitory effect of MEG3/miR-214/AIFM2 axis on the growth of T-cell lymphoblastic lymphoma Retraction. Int J Oncol. 2017;51(1):316–26. https://doi.org/10.3892/ijo.2020.5162.
Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–91.
Su L, Kong H, Wu F, Lv H, Wu W, Wang G, et al. Long non-coding RNA zinc finger antisense 1 functions as an oncogene in acute promyelocytic leukemia cells. Oncol Lett. 2019;18(6):6331–8.
Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood J Am Soc Hematol. 2011;117(26):7053–62.
Swindall AF, Londoño-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines ST6Gal-I is upregulated in epithelial cancers and CSCs. Can Res. 2013;73(7):2368–78.
Zhang B, Sun Y, Zhang X, Jiang N, Chen Q. TUG1 weakens the sensitivity of acute myeloid leukemia cells to cytarabine by regulating miR-655-3p/CCND1 axis. Eur Rev Med Pharmacol Sci. 2020;24(9):4940–53.
Dai S, Li N, Zhou M, Yuan Y, Yue D, Li T, et al. LncRNA EBLN3P promotes the progression of osteosarcoma through modifying the miR-224-5p/Rab10 signaling axis. Sci Rep. 2021;11(1):1–12.
Mathias C, Muzzi JCD, Antunes BB, Gradia DF, Castro MA, Carvalho de Oliveira J. Unraveling immune-related lncRNAs in breast cancer molecular subtypes. Front Oncol 2021;11:692170.
Gao W. Long non-coding RNA MEG3 as a candidate prognostic factor for induction therapy response and survival profile in childhood acute lymphoblastic leukemia patients. Scand J Clin Lab Invest. 2021;81(3):194–200.
Pei J-S, Chang W-S, Chen C-C, Mong M-C, Hsu S-W, Hsu P-C, et al. Novel contribution of long non-coding RNA MEG3 genotype to prediction of childhood leukemia risk. Cancer Genom Proteom. 2022;19(1):27–34.
Li Q, Wang J. Long noncoding RNA ZFAS1 enhances adriamycin resistance in pediatric acute myeloid leukemia through the miR-195/Myb axis. RSC Adv. 2019;9(48):28126–34.
Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–8.
Song Z, Wu W, Chen M, Cheng W, Yu J, Fang J, et al. Long noncoding RNA ANRIL supports proliferation of adult T-cell leukemia cells through cooperation with EZH2. J Virol. 2018;92(24):e00909-e918.
Chen L, Shi Y, Li J, Yang X, Li R, Zhou X, et al. LncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335–3p/TRAF5 axis. Anticancer Drugs. 2020. https://doi.org/10.1097/CAD.0000000000001001.
Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66.
Zhang E-B, Kong R, Yin D-D, You L-H, Sun M, Han L, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276.
Zhou R, Mo W, Wang S, Zhou W, Chen X, Pan S. miR-141–3p and TRAF5 network contributes to the progression of T-cell acute lymphoblastic leukemia. Cell Transplant. 2019;28(1_suppl):59S-65S.
Author information
Authors and Affiliations
Contributions
HY, BK and EL designed the study and supervised the data collection, DP, MJ, and M-RM-S prepared the manuscript for publication and reviewed the draft of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing of interest.
Conflict of interests
Healthy and patient people did not participate in this study and therefore there is no need for consent.
Ethical approval and consent to participate
Not applicable
Consent for publication
Not applicable
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yousefi, H., Purrahman, D., Jamshidi, M. et al. Long non-coding RNA signatures and related signaling pathway in T-cell acute lymphoblastic leukemia. Clin Transl Oncol 24, 2081–2089 (2022). https://doi.org/10.1007/s12094-022-02886-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02886-9