Abstract
Purpose
Metabolomics is an emerging field in cancer research. Plasma free amino acid profiles (PFAAs) have shown different features in various cancers, but the characteristic in advanced sarcoma remains unclear. We aimed to uncover the specific PFAAs in advanced sarcoma and to find the relationship between the altering of PFAAs and response to chemotherapy.
Patients and methods
We analyzed the differences in PFAAs between 23 sarcoma patients and 30 healthy subjects basing on liquid chromatography–tandem mass spectrometry (LC–MS/MS). Then, we compared the dynamics of PFAAs after chemotherapy between improvement group and deterioration group.
Results
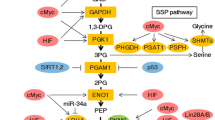
We identified seven biological differential amino acids and four pathways which were perturbed in the sarcoma patients compared with healthy subjects. After one cycle chemotherapy, the levels of γ-aminobutyric acid (GABA) and carnosine (Car) decreased significantly in the improvement group but not in deterioration group. The levels of α-aminobutyric acid (Abu) increased significantly in the deterioration group but not in the improvement group.
Conclusion
Our study suggests the potential specific PFAAs in sarcoma patients. The unusual amino acids and metabolic pathways may provide ideas for clinical drugs targeting therapy. Three amino acids including Car, GABA and Abu may be metabolic biomarkers playing a role in dynamic monitoring of the therapeutic effect.




Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30.
Burningham Z, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res. 2012;2:14.
Bastiaannet E, Groen H, Jager PL, Cobben DC, van der Graaf WT, Vaalburg W, Hoekstra HJ. The value of FDG-PET in the detection, grading and response to therapy of soft tissue and bone sarcomas; a systematic review and meta-analysis. Cancer Treat Rev. 2004;30:83–101.
Schaefer IM, Cote GM, Hornick JL. Contemporary sarcoma diagnosis, genetics, and genomics. J Clin Oncol. 2018;36:101–10.
Skubitz KM, D’Adamo DR. Sarcoma. Mayo Clin Proc. 2007;82:1409–32.
Pisters PW, Leung DH, Woodruff J, Shi W, Brennan MF. Analysis of prognostic factors in 1,041 patients with localized soft tissue sarcomas of the extremities. J Clin Oncol. 1996;14:1679–89.
Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3:685–94.
Dancsok AR, Asleh-Aburaya K, Nielsen TO. Advances in sarcoma diagnostics and treatment. Oncotarget. 2017;8:7068–93.
Molina-Ortiz D, Torres-Zarate C, Cardenas-Cardos R, Palacios-Acosta JM, Hernandez-Arrazola D, Shalkow-Klincovstein J, Diaz-Diaz E, Vences-Mejia A. MDR1 not CYP3A4 gene expression is the predominant mechanism of innate drug resistance in pediatric soft tissue sarcoma patients. Cancer Biomark. 2018;22:317–24.
Leichtle AB, Nuoffer JM, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8:643–53.
Proenza AM, Oliver J, Palou A, Roca P. Breast and lung cancer are associated with a decrease in blood cell amino acid content. J Nutr Biochem. 2003;14:133–8.
Lai HS, Lee JC, Lee PH, Wang ST, Chen WJ. Plasma free amino acid profile in cancer patients. Semin Cancer Biol. 2005;15:267–76.
Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, Saruki N, Bando E, Kimura H, Imamura F, Moriyama M, Ikeda I, Chiba A, Oshita F, Imaizumi A, Yamamoto H, Miyano H, Horimoto K, Tochikubo O, Mitsushima T, Yamakado M, Okamoto N. Plasma free amino acid profiling of five types of cancer patients and its application for early detectio. PLoS ONE. 2011;6:e24143.
Gu Y, Chen T, Fu S, Sun X, Wang L, Wang J, Lu Y, Ding S, Ruan G, Teng L, Wang M. Perioperative dynamics and significance of amino acid profiles in patients with cancer. J Transl Med. 2015;13:35.
Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–60.
Harder U, Koletzko B, Peissner W. Quantification of 22 plasma amino acids combining derivatization and ion-pair LC-MS/MS. J Chromatogr B. 2011;879:495–504.
Kubota A, Meguid MM, Hitch DC. Amino acid profiles correlate diagnostically with organ site in three kinds of malignant tumors. Cancer. 1992;69:2343–8.
Clarke EF, Lewis AM, Waterhouse C. Peripheral amino acid levels in patients with cancer. Cancer. 1978;42:2909–13.
Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistanc. Cell Metab. 2009;9:311–26.
Nie C, He T, Zhang W, Zhang G, Ma X. Branched chain amino acids: beyond nutrition metabolis. Int J Mol Sci. 2018;19:954.
Lee JH, Cho YR, Kim JH, Kim J, Nam HY, Kim SW, Son J. Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Exp Mol Med. 2019;51:1–11.
Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, Edison AS, Ito T. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545:500–4.
Fischer JE, Chance WT. Total parenteral nutrition, glutamine, and tumor growth. JPEN J Parenter Enteral Nutr. 1990;14:86S–9S.
Norton JA, Gorschboth CM, Wesley RA, Burt ME, Brennan MF. Fasting plasma amino acid levels in cancer patient. Cancer. 1985;56:1181–6.
Min L, Choy E, Tu C, Hornicek F, Duan Z. Application of metabolomics in sarcoma: from biomarkers to therapeutic targets. Crit Rev Oncol Hematol. 2017;116:1–10.
Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: opportunities and challenges. Cancer Res Treat. 2013;45:251–62.
Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur J Biochem. 2003;270:1887–99.
Dillon BJ, Prieto VG, Curley SA, Curley A, Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer. 2004;100:826–33.
Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, Morioka H, Toyama Y, Hirohashi S, Kawai A, Yamada T. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–44.
Huang HY, Wu WR, Wang YH, Wang JW, Fang FM, Tsai JW, Li SH, Hung HC, Yu SC, Lan J, Shiue YL, Hsing CH, Chen LT, Li CF. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin Cancer Res. 2013;19:2861–72.
Closs EI, Scheld JS, Sharafi M, Forstermann U. Substrate supply for nitric-oxide synthase in macrophages and endothelial cells: role of cationic amino acid transporters. Mol Pharmacol. 2000;57:68–74.
Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4.
Yoon JK, Kim DH, Koo JS. Implications of differences in expression of sarcosine metabolism-related proteins according to the molecular subtype of breast cancer. J Transl Med. 2014;12:149.
Jentzmik F, Stephan C, Lein M, Miller K, Kamlage B, Bethan B, Kristiansen G, Jung K. Sarcosine in prostate cancer tissue is not a differential metabolite for prostate cancer aggressiveness and biochemical progression. J Urol. 2011;185:706–11.
Shajahan-Haq A, Cheema M, Clarke R. Application of metabolomics in drug resistant breast cancer research. Metabolites. 2015;5:100–18.
Al-Zoughbi W, Huang J, Paramasivan GS, Till H, Pichler M, Guertl-Lackner B, Hoefler G. Tumor macroenvironment and metabolism. Semin Oncol. 2014;41:281–95.
Tan GG, Zhao BB, Li YQ, Liu X, Zou ZL, Wan J, Yao Y, Xiong H, Wang YY. Pharmacometabolomics identifies dodecanamide and leukotriene B4 dimethylamide as a predictor of chemosensitivity for patients with acute myeloid leukemia treated with cytarabine and anthracycline. Oncotarget. 2017;8:88697–707.
Man TK, Chintagumpala M, Visvanathan J, Shen J, Perlaky L, Hicks J, Johnson M, Davino N, Murray J, Helman L, Meyer W, Triche T, Wong KK, Lau CC. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res. 2005;65:8142–50.
Boldyrev AA, Aldini G, Derave W. Physiology and pathophysiology of carnosine. Physiol Rev. 2013;93:1803–45.
Gaunitz F, Hipkiss AR. Carnosine and cancer: a perspective. Amino Acids. 2012;43:135–42.
Zhang Z, Miao L, Wu X, Liu G, Peng Y, Xin X, Jiao B, Kong X. Carnosine inhibits the proliferation of human gastric carcinoma cells by retarding Akt/mTOR/p70S6K signaling. J Cancer. 2014;5:382–9.
Bao Y, Ding S, Cheng J, Liu Y, Wang B, Xu H, Shen Y, Lyu J. Carnosine inhibits the proliferation of human cervical gland carcinoma cells through inhibiting both mitochondrial bioenergetics and glycolysis pathways and retarding cell cycle progression. Integr Cancer Ther. 2018;17:80–91.
Hwang B, Shin SS, Song JH, Choi YH, Kim WJ, Moon SK. Carnosine exerts antitumor activity against bladder cancers in vitro and in vivo via suppression of angiogenesis. J Nutr Biochem. 2019;74:108230.
Andersson AC, Henningsson S, Jarhult J. Diamine oxidase activity and gamma-aminobutyric acid formation in medullary carcinoma of the thyroid. Agents Actions. 1980;10:299–301.
Nicholson-Guthrie CS, Guthrie GD, Sutton GP, Baenziger JC. Urine GABA levels in ovarian cancer patients: elevated GABA in malignancy. Cancer Lett. 2001;162:27–30.
Kleinrok Z, Matuszek M, Jesipowicz J, Matuszek B, Opolski A, Radzikowski C. GABA content and GAD activity in colon tumors taken from patients with colon cancer or from xenografted human colon cancer cells growing as s.c. tumors in athymic nu/nu mice. J Physiol Pharmacol. 1998;49:303–10.
Mazurkiewicz M, Opolski A, Wietrzyk J, Radzikowski C, Kleinrok Z. GABA level and GAD activity in human and mouse normal and neoplastic mammary gland. J Exp Clin Cancer Res. 1999;18:247–53.
Joseph J, Niggemann B, Zaenker KS, Entschladen F. The neurotransmitter gamma-aminobutyric acid is an inhibitory regulator for the migration of SW 480 colon carcinoma cells. Cancer Res. 2002;62:6467–9.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81872264).
Author information
Authors and Affiliations
Contributions
YQ was responsible for conception and supervision. BJ and WW prepared the manuscript. WW, SL, YL and LS interpreted and analyzed data. BJ revised the important intellectual content. YG and FG provided technical support for plasma testing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical standards
The content and processes of the study have been reviewed and approved by the Ethics Committee of Zhengzhou University which is guided by international and national ethical requirements. In compliance with the Declaration of Helsinki, patient data were maintained with strict confidentiality.
Informed consent
Patients were informed in advance before taking blood samples, and all patients were informed of the treatment process and signed informed consent documents.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jia, B., Wang, W., Lin, S. et al. The free amino acid profiles and metabolic biomarkers of predicting the chemotherapeutic response in advanced sarcoma patients. Clin Transl Oncol 22, 2213–2221 (2020). https://doi.org/10.1007/s12094-020-02494-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-020-02494-5