Abstract
Background
Acute myeloid leukemia (AML) is the most common type of acute leukemia and biologically heterogeneous diseases with poor prognosis. Thus, we aimed to identify prognostic markers to effectively predict the prognosis of AML patients and eventually guide treatment.
Methods
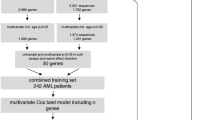
Prognosis-associated genes were determined by Kaplan–Meier and multivariate analyses using the expression and clinical data of 173 AML patients from The Cancer Genome Atlas database and validated in an independent Oregon Health and Science University dataset. A prognostic risk score was computed based on a linear combination of 5-gene expression levels using the regression coefficients derived from the multivariate logistic regression model. The classification of AML was established by unsupervised hierarchical clustering of CALCRL, DOCK1, PLA2G4A, FCHO2 and LRCH4 expression levels.
Results
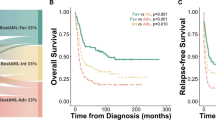
High FCHO2 and LRCH4 expression was related to decreased mortality. While high CALCRL, DOCK1, PLA2G4A expression was associated with increased mortality. The risk score was predictive of increased mortality rate in AML patients. Hierarchical clustering analysis of the five genes discovered three clusters of AML patients. The cluster1 AML patients were associated with lower cytogenetics risk than cluster2 or 3 patients, and better prognosis than cluster3 patients (P values < 0.05 for all cases, fisher exact test or log-rank test).
Conclusion
The gene panel comprising CALCRL, DOCK1, PLA2G4A, FCHO2 and LRCH4 as well as the risk score may offer novel prognostic biomarkers and classification of AML patients to significantly improve outcome prediction.




Similar content being viewed by others
Availability of data and material
The datasets generated and/or analysed during the current study are available upon reasonable request.
References
Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–907.
Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:29. https://onlinelibrary.wiley.com/doi/10.3322/caac.21254/pdf
Shah A, Andersson TML, Rachet B, Björkholm M, Lambert PC. Survival and cure of acute myeloid leukaemia in England, 1971–2006: a population-based study. Br J Haematol. 2013;162:509–16.
Meyers J, Yu Y, Kaye JA, Davis KL. Medicare fee-for-service enrollees with primary acute myeloid leukemia: an analysis of treatment patterns, survival, and healthcare resource utilization and costs. Appl Health Econ Health Policy. 2013;11:275–86. https://doi.org/10.1007/s40258-013-0032-2.
Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–36.
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009;106:12950–5. https://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2716381&tool=pmcentrez&rendertype=abstract
Bullinger L, Krönke J, Schön C, Radtke I, Urlbauer K, Botzenhardt U, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010;24:438–49.
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. https://doi.org/10.1056/NEJMoa1112304 (Massachusetts Medical Society).
Network TCGAR. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med. 2013;368:2059–74. https://www.nejm.org/doi/10.1056/NEJMoa1301689
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31. https://doi.org/10.1038/s41586-018-0623-z.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1–pl1. https://pubmed.ncbi.nlm.nih.gov/23550210
Therneau T. Survival Analysis. Cran [Internet]. 2016; https://cran.r-project.org/web/packages/survival/survival.pdf
Fox J. Cox Proportional-Hazards Regression for Survival Data The Cox Proportional-Hazards Model. Most. 2002;2008:1–18. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.110.2264&rep=rep1&type=pdf
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. BioMed Central Ltd; 2011;12:77. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-77
Consortium Gte. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. https://pubmed.ncbi.nlm.nih.gov/23715323
Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. gplots: Various R programming tools for plotting data. R Packag. version. 2005.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. https://pubmed.ncbi.nlm.nih.gov/27895058 (American Society of Hematology)
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Ciftciler R, Goker H, Buyukasık Y, Malkan UY, Saglam EA. Impact of pre-transplant bone marrow blast percentage on survival in acute myeloid leukemia patients. Int J Clin Exp Med. 2019;12:9288–94.
Lee S, Chiu Y, Li Y, Lin C, Hou H, Chou W, et al. High Expression of dedicator of Cytokinesis 1 ( DOCK1) Confers Poor Prognosis in Acute Myeloid Leukemia. Oncotarget. 2017;8:72250–9.
Angenendt L, Bormann E, Pabst C, Alla V, Görlich D, Braun L, et al. The neuropeptide receptor calcitonin receptor-like (CALCRL) is a potential therapeutic target in acute myeloid leukemia. Leukemia. 2019;33:2830–41. https://doi.org/10.1038/s41375-019-0505-x.
Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. https://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gkl842
Yang L, Zhang H. Expression of cytosolic phospholipase A2 alpha in glioblastoma is associated with resistance to chemotherapy. Am J Med Sci. 2018;356:391–8. https://doi.org/10.1016/j.amjms.2018.06.019.
Runarsson G, Feltenmark S, Forsell PKA, Sjöberg J, Björkholm M, Claesson H-E. The expression of cytosolic phospholipase A2 and biosynthesis of leukotriene B4 in acute myeloid leukemia cells. Eur J Haematol. 2007;79:468–76. https://doi.org/10.1111/j.1600-0609.2007.00967.x.
Sundarraj S, Kannan S, Thangam R, Gunasekaran P. Effects of the inhibition of cytosolic phospholipase A2α in non-small cell lung cancer cells. J Cancer Res Clin Oncol. 2012;138:827–35. https://doi.org/10.1007/s00432-012-1157-7.
Parhamifar L, Jeppsson B, Sjölander A. Activation of cPLA2 is required for leukotriene D4-induced proliferation in colon cancer cells. Carcinogenesis. 2005;26:1988–98.
Tajiri H, Uruno T, Shirai T, Takaya D, Matsunaga S, Setoyama D, et al. Targeting ras-driven cancer cell survival and invasion through selective inhibition of DOCK1. Cell Rep. 2017;19:969–80.
Funding
None.
Author information
Authors and Affiliations
Contributions
KS designed the study. RP, KS, LC downloaded somatic mutation, RNA-seq data and clinical data from the TCGA and OHSU databases. YL, PZ, RP, XS performed the survival analyses and differentially expressed gene analysis. ZF, LC conducted unsupervised hierarchical clustering analysis. KS prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
The study was approved by the Ethics Committee of the Department and conformed to the ethical guidelines of the Helsinki Declaration (as revised in Tykyo 2004).
Informed consent
All participants have provided written informed consent in this study.
Code availability
The codes in the current study are available upon reasonable request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12094_2020_2460_MOESM2_ESM.tif
Supplementary file2 Supplementary Figure 1. The overlap of prognosis-associated genes between TCGA and OHSU datasets. A. The overlap of protective type genes between TCGA and OHSU datasets, A. The overlap of risk type genes between TCGA and OHSU datasets. (TIF 1959 kb)
12094_2020_2460_MOESM3_ESM.tif
Supplementary file3 Supplementary Figure 2. Kaplan-Meier survival analysis of patients’ OS with CALCRL (A), DOCK1 (B), FCHO2 (C) and LRCH4 (D) and PLA2G4A (E) expression levels in 405 AML patients of the OHSU dataset. (TIF 1232 kb)
12094_2020_2460_MOESM4_ESM.tif
Supplementary file4 Supplementary Figure 3. The mutation frequencies of DOCK1 (A), FCHO2 (B) and PLA2G4A (C) in the TCGA and OHSU datasets. (TIF 1898 kb)

12094_2020_2460_MOESM5_ESM.jpg
Supplementary file5 Supplementary Figure 4. Unsupervised hierarchical clustering of the 5-gene panel uncovered three classes of AML patients in the TCGA dataset. (JPEG 1663 kb)

12094_2020_2460_MOESM6_ESM.jpg
Supplementary file6 Supplementary Figure 5. Unsupervised hierarchical clustering of the 5-gene panel uncovered three classes of AML patients in the OHSU dataset. (JPEG 978 kb)
12094_2020_2460_MOESM7_ESM.tif
Supplementary file7 Supplementary Figure 6. The three clusters of AML patients exhibited significant differences in FLT3-ITD mutation in the OHSU dataset. (TIF 1847 kb)
Rights and permissions
About this article
Cite this article
Sha, K., Lu, Y., Zhang, P. et al. Identifying a novel 5-gene signature predicting clinical outcomes in acute myeloid leukemia. Clin Transl Oncol 23, 648–656 (2021). https://doi.org/10.1007/s12094-020-02460-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-020-02460-1