Abstract
Purpose
The prognosis of AML patients with chemotherapy is poor, especially those who are insensitive to and resistant to chemotherapy drugs. To clarify the underlying pathogenesis of AML and provide new therapeutic targets for clinical treatment, we explore the role of circRNA in leukemia.
Methods
High-throughput circRNA sequencing analysis was performed in patients with leukemia and healthy donors. RT-qPCR and western blot analysis were used to determine expression of GSK3β. RNA pull-down assay was used to detect miRNAs pulled down by hsa_circ_0121582. RNA immunoprecipitation assay was performed to evaluate the binding capacity between TET1 and hsa_circ_0121582.
Results
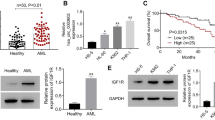
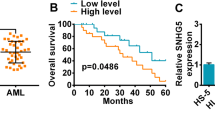
A new and highly stable circRNA was found, which was derived from the reverse splicing of GSK3β exon 1 to exon 7, and hsa_circ_0121582 was down-regulated in leukemia cells. In gain-of-function experiments, the up-regulated hsa_circ_0121582 inhibited the proliferation of leukemia cells in vitro and in vivo. In the cytoplasm, hsa_circ_0121582 could act as a sponge for miR-224, attenuate the inhibiting effect of miR-224 on GSK3β, and thus up-regulate the expression level of GSK3β. In addition, hsa_circ_0121582 could bind to GSK3β promoter in the nucleus, and recruit DNA demethylase TET1 to ensuring the transcription of GSK3β. The upregulated GSK3β inhibited the Wnt/β-catenin signaling pathway, and reduced the aggregation of β-catenin in the nucleus, thus inhibited the proliferation of leukemia cells.
Conclusions
This study found that hsa_circ_0121582 was involved in the inhibition of tumor proliferation, and the restoration of hsa_circ_0121582 could be an effective treatment strategy for patients with leukemia.




Similar content being viewed by others
References
Abdul-Aziz AM, Sun Y, Hellmich C, Marlein CR, Mistry J, Forde E, et al. Acute myeloid leukemia induces protumoral p16INK4a-driven senescence in the bone marrow microenvironment. Blood. 2019;133:446–56.
Shallis RM, Boddu PC, Bewersdorf JP, Zeidan AM. The golden age for patients in their golden years: The progressive upheaval of age and the treatment of newly-diagnosed acute myeloid leukemia. Blood Rev. 2019;2019:100639.
Chen X, Pan J, Wang S, Hong S, Hong S, He S. The epidemiological trend of acute myeloid leukemia in childhood: a population-based analysis. J Cancer. 2019;10:4824–35.
Li G, Zhou Z, Yang W, Yang H, Fan X, Yin Y, et al. Long-term cardiac-specific mortality among 44,292 acute myeloid leukemia patients treated with chemotherapy: a population-based analysis. J Cancer. 2019;10:6161–9.
Patel HP, Perissinotti AJ, Patel TS, Bixby DL, Marshall VD, Marini BL. Incidence and risk factors for breakthrough invasive mold infections in acute myeloid leukemia patients receiving remission induction chemotherapy. Open Forum Infect Dis. 2019;6:176.
Ricci A, Jin Z, Broglie L, Bhatia M, George D, Garvin JH, et al. Healthcare utilization and financial impact of acute-graft-versus host disease among children undergoing allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2019;55(2):384.
Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med. 2019;70:141–52.
Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630–8.
Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 2019;11:55.
Cao S, Ma T, Ungerleider N, Roberts C, Kobelski M, Jin L, et al. Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene. 2019;38:7060–72.
Kolling M, Haddad G, Wegmann U, Kistler A, Bosakova A, Seeger H, et al. Circular RNAs in urine of kidney transplant patients with acute T cell-mediated allograft rejection. Clin Chem. 2019;65:1287–94.
Wu Z, Sun H, Li J, Jin H. Circular RNAs in leukemia. Aging. 2019;11:4757–71.
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–40.
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. ncx1A circular transcript of gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–69.
Ma N, Pan J, Ye X, Yu B, Zhang W, Wan J. Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol Ther Nucleic Acids. 2019;18:1049–62.
Wang G, Guo X, Cheng L, Chu P, Chen M, Chen Y, et al. An integrated analysis of the circRNA-miRNA-mRNA network reveals novel insights into potential mechanisms of cell proliferation during liver regeneration. Artif Cells Nanomed Biotechnol. 2019;47:3873–84.
Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015–21.
Ma X, Liu C, Gao C, Li J, Zhuang J, Liu L, et al. circRNA-associated ceRNA network construction reveals the circRNAs involved in the progression and prognosis of breast cancer. J Cell Physiol. 2019;235(4):3973.
Li HM, Dai YW, Yu JY, Duan P, Ma XL, Dong WW, et al. Comprehensive circRNA/miRNA/mRNA analysis reveals circRNAs protect against toxicity induced by BPA in GC-2 cells. Epigenomics. 2019;11:935–49.
Raveendra BL, Swarnkar S, Avchalumov Y, Liu XA, Grinman E, Badal K, et al. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc Natl Acad Sci USA. 2018;115:E10197–E1020510205.
Lu G, Zhang J, Liu X, Liu W, Cao G, Lv C, et al. Regulatory network of two circRNAs and an miRNA with their targeted genes under astilbin treatment in pulmonary fibrosis. J Cell Mol Med. 2019;23:6720–9.
Wang J, Yin J, Wang X, Liu H, Hu Y, Yan X, et al. Changing expression profiles of mRNA, lncRNA, circRNA, and miRNA in lung tissue reveal the pathophysiological of bronchopulmonary dysplasia (BPD) in mouse model. J Cell Biochem. 2019;120:9369–80.
Author information
Authors and Affiliations
Contributions
All authors have participated in the analysis and interpretation of data, drafting of the first version of the manuscript, revising critically subsequent versions, and approving the fnal version of the manuscript before submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interests to declare.
Ethical approval
All patients involved consented to participate under the Clinical Patient ethical statement. The study was approved by the Institutional Ethical Board of The First-Affiliated Hospital of Hunan Normal University.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, JJ., Lei, P. & Zhou, M. hsa_circ_0121582 inhibits leukemia growth by dampening Wnt/β-catenin signaling. Clin Transl Oncol 22, 2293–2302 (2020). https://doi.org/10.1007/s12094-020-02377-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-020-02377-9