Abstract
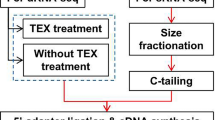
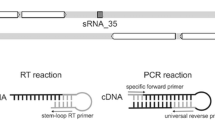
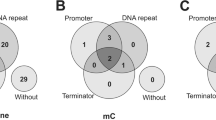
Regulatory small RNAs (sRNA) are RNA transcripts that are not translated into proteins but act as functional RNAs. Pathogenic Leptospira cause an epidemic spirochaetal zoonosis, Leptospirosis. It is speculated that Leptospiral sRNAs are involved in orchestrating their pathogenicity. In this study, biocomputational approach was adopted to identify Leptospiral sRNAs. In this study, two sRNA prediction programs, i.e., RNAz and nocoRNAc, were employed to screen the reference genome of Leptospira interrogans serovar Lai. Out of 126 predicted sRNAs, there are 96 cis-antisense sRNAs, 28 trans-encoded sRNAs and 2 sRNAs that partially overlap with protein-coding genes in a sense orientation. To determine whether these candidates are expressed in the pathogen, they were compared with the coverage files generated from our RNA-seq datasets. It was found out that 7 predicted sRNAs are expressed in mid-log phase, stationary phase, serum stress, temperature stress and iron stress while 2 sRNAs are expressed in mid-log phase, stationary phase, serum stress, and temperature stress. Besides, their expressions were also confirmed experimentally via RT-PCR. These experimentally validated candidates were also subjected to mRNA target prediction using TargetRNA2. Taken together, our study demonstrated that biocomputational strategy can serve as an alternative or as a complementary strategy to the laborious and expensive deep sequencing methods not only to uncover putative sRNAs but also to predict their targets in bacteria. In fact, this is the first study that integrates computational approach to predict putative sRNAs in L. interrogans serovar Lai.


Similar content being viewed by others
References
Cheah H-L, Raabe CA, Lee L-P, Rozhdestvensky TS, Citartan M, Ahmed SA, Tang T-H (2018) Bacterial regulatory RNAs: complexity, function, and putative drug targeting. Crit Rev Biochem Mol Biol 53:335–355. https://doi.org/10.1080/10409238.2018.1473330
Novick RP, Ross H, Projan S, Kornblum J, Kreiswirth B, Moghazeh S (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975
Viegas SC, Arraiano CM (2008) Regulating the regulators: how ribonucleases dictate the rules in the control of small non-coding RNAs. RNA Biol 5:230–243. https://doi.org/10.4161/rna.6915
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. https://doi.org/10.1038/nature02871
Babski J, Maier L-K, Heyer R, Jaschinski K, Prasse D, Jäger D, Randau L, Schmitz RA, Marchfelder A, Soppa J (2014) Small regulatory RNAs in Archaea. RNA Biol 11:484–493. https://doi.org/10.4161/rna.28452
Hüttenhofer A, Brosius J, Bachellerie JP (2002) RNomics: identification and function of small, non-messenger RNAs. Curr Opin Chem Biol 6:835–843. https://doi.org/10.1016/s1367-5931(02)00397-6
Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16. https://doi.org/10.1016/j.ydbio.2005.10.036
Hoe C-H, Raabe CA, Rozhdestvensky TS, Tang T-H (2013) Bacterial sRNAs: regulation in stress. Int J Med Microbiol 303:217–229
Morfeldt E, Dv T, Von Gabain A, Arvidson S (1995) Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14:4569–4577
Paulander W, Varming AN, Bojer MS, Friberg C, Bæk K, Ingmer H (2018) The agr quorum sensing system in Staphylococcus aureus cells mediates death of sub-population. BMC Res Notes 11:503. https://doi.org/10.1186/s13104-018-3600-6
Gupta RK, Luong TT, Lee CY (2015) RNAIII of the <em>Staphylococcus aureus agr</em> system activates global regulator MgrA by stabilizing mRNA. Proc Natl Acad Sci 112:14036–14041. https://doi.org/10.1073/pnas.1509251112
Mohammed H, Nozha C, Hakim K, Abdelaziz F, Rekia B (2011) Leptospira: morphology, classification and pathogenesis. J Bacteriol Parasitol. https://doi.org/10.4172/2155-9597.1000120
Bandara M, Ananda M, Wickramage K, Berger E, Agampodi S (2014) Globalization of leptospirosis through travel and migration. Glob Health 10:61
Crawford R, Heinemann J, McCulloch W, Diesch S (1971) Human infections associated with waterborne Leptospires, and survival studies on serotype pomona. J Am Vet Med Assoc 159:1477–1484
Jaksik R, Iwanaszko M, Rzeszowska-Wolny J, Kimmel M (2015) Microarray experiments and factors which affect their reliability. Biol Direct 10:46
Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T (2010) Visualizing genomes: techniques and challenges. Nat Methods 7:S5
Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP (2010) Computational solutions to large-scale data management and analysis. Nat Rev Genet 11:647
Washietl S, Hofacker IL, Stadler PF (2005) Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci USA 102:2454–2459. https://doi.org/10.1073/pnas.0409169102
Gruber AR, Findeiß S, Washietl S, Hofacker IL, Stadler PF (2010) RNAz 2.0: improved noncoding RNA detection. Pacific Symposium on Biocomputing, pp 69–79
Herbig A, Nieselt K (2011) nocoRNAc: characterization of non-coding RNAs in prokaryotes. BMC Bioinform 12:40. https://doi.org/10.1186/1471-2105-12-40
Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. https://doi.org/10.1101/gr.2289704
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Kingsford CL, Ayanbule K, Salzberg SL (2007) Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol 8:R22. https://doi.org/10.1186/gb-2007-8-2-r22
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR (2003) Rfam: an RNA family database. Nucleic Acids Res 31:439–441. https://doi.org/10.1093/nar/gkg006
Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI (2018) Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinform 62:e51. https://doi.org/10.1002/cpbi.51
Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, Marz M, Griffiths-Jones S, Toffano-Nioche C, Gautheret D, Weinberg Z, Rivas E, Eddy SR, Finn Robert D, Bateman A, Petrov AI (2020) Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res 49:D192–D200. https://doi.org/10.1093/nar/gkaa1047
Hertel J, de Jong D, Marz M, Rose D, Tafer H, Tanzer A, Schierwater B, Stadler PF (2009) Non-coding RNA annotation of the genome of Trichoplax adhaerens. Nucleic Acids Res 37:1602–1615. https://doi.org/10.1093/nar/gkn1084
Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R (2007) Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol 3:e65. https://doi.org/10.1371/journal.pcbi.0030065
Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R (2012) LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA 18:900–914. https://doi.org/10.1261/rna.029041.111
Raden M, Ali SM, Alkhnbashi OS, Busch A, Costa F, Davis JA, Eggenhofer F, Gelhausen R, Georg J, Heyne S, Hiller M, Kundu K, Kleinkauf R, Lott SC, Mohamed MM, Mattheis A, Miladi M, Richter AS, Will S, Wolff J, Wright PR, Backofen R (2018) Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Res 46:W25-w29. https://doi.org/10.1093/nar/gky329
Kery MB, Feldman M, Livny J, Tjaden B (2014) TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res 42:W124–W129. https://doi.org/10.1093/nar/gku317
Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, Kleinkauf R, Hess WR, Backofen R (2014) CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res 42:W119–W123. https://doi.org/10.1093/nar/gku359
Pánek J, Bobek J, Mikulík K, Basler M, Vohradský J (2008) Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics 9:217. https://doi.org/10.1186/1471-2164-9-217
Rivas E, Klein RJ, Jones TA, Eddy SR (2001) Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol 11:1369–1373. https://doi.org/10.1016/s0960-9822(01)00401-8
Rossi CC, Bossé JT, Li Y, Witney AA, Gould KA, Langford PR, Bazzolli DMS (2016) A computational strategy for the search of regulatory small RNAs in Actinobacillus pleuropneumoniae. RNA 22:1373–1385. https://doi.org/10.1261/rna.055129.115
Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR (2005) Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J 24:3360–3368. https://doi.org/10.1038/sj.emboj.7600805
Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR (2002) Genetic control by a metabolite binding mRNA. Chem Biol 9:1043–1049. https://doi.org/10.1016/S1074-5521(02)00224-7
Guo S, Frazer DM, Anderson GJ (2016) Iron homeostasis: transport, metabolism, and regulation. Curr Opin Clin Nutr Metab Care 19:276–281. https://doi.org/10.1097/mco.0000000000000285
Harris ED (2000) Cellular copper transport and metabolism. Annu Rev Nutr 20:291–310. https://doi.org/10.1146/annurev.nutr.20.1.291
Udekwu KI, Darfeuille F, Vogel J, Reimegård J, Holmqvist E, Wagner EGH (2005) Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19:2355–2366. https://doi.org/10.1101/gad.354405
Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U (2000) Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev 14:1109–1118
Updegrove TB, Shabalina SA, Storz G (2015) How do base-pairing small RNAs evolve? FEMS Microbiol Rev 39:379–391
Zhukova A, Fernandes LG, Hugon P, Pappas CJ, Sismeiro O, Coppée JY, Becavin C, Malabat C, Eshghi A, Zhang JJ, Yang FX, Picardeau M (2017) Genome-Wide Transcriptional Start Site Mapping and sRNA Identification in the Pathogen Leptospira interrogans. Front Cell Infect Microbiol 7:10. https://doi.org/10.3389/fcimb.2017.00010
Chheda N, Gupta MK (2014) RNA as a permutation. arXiv preprint arXiv:1403.5477
Messias AC, Sattler M (2004) Structural basis of single-stranded RNA recognition. Acc Chem Res 37:279–287
Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL (2006) Thermodynamics of RNA–RNA binding. Bioinformatics 22:1177–1182
McCullen CA, Benhammou JN, Majdalani N, Gottesman S (2010) Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 192:5559–5571
Orelle C, Mathieu K, Jault JM (2019) Multidrug ABC transporters in bacteria. Res Microbiol 170:381–391. https://doi.org/10.1016/j.resmic.2019.06.001
Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F, Stulik J (2013) Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun 81:629–635. https://doi.org/10.1128/iai.01035-12
Bröms JE, Edqvist PJ, Forsberg A, Francis MS (2006) Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol Lett 256:57–66. https://doi.org/10.1111/j.1574-6968.2005.00099.x
Edqvist PJ, Bröms JE, Betts HJ, Forsberg A, Pallen MJ, Francis MS (2006) Tetratricopeptide repeats in the type III secretion chaperone, LcrH: their role in substrate binding and secretion. Mol Microbiol 59:31–44. https://doi.org/10.1111/j.1365-2958.2005.04923.x
Chakraborty S, Monfett M, Maier TM, Benach JL, Frank DW, Thanassi DG (2008) Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect Immun 76:2852–2861. https://doi.org/10.1128/iai.01726-07
Chao J, Wong D, Zheng X, Poirier V, Bach H, Hmama Z, Av-Gay Y (2010) Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochem Biophys Acta 1804:620–627. https://doi.org/10.1016/j.bbapap.2009.09.008
Acknowledgements
This project was supported by FRGS grant (Grant #: 203/CIPPT/6711510) from the Ministry of Higher Education (MOHE), Malaysia, which was awarded to THT. SA holds an e-ScienceFund (Grant #:305/CIPPT/613237) from the Ministry of Science, Malaysia. XYT was supported by MyBrain15 program (KPT(B) 930521075093) under Malaysian Ministry of Education.
Funding
FRGS, 203/CIPPT/6711510, Tang Thean Hock, E-science, 05/CIPPT/613237, Siti Aminah Ahmad, MyBrain15 program, KPT(B) 930521075093, Xinq Yuan Tan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interest with respect to the authorship and/or publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tan, X.Y., Citartan, M., Chinni, S.V. et al. Biocomputational Identification of sRNAs in Leptospira interrogans Serovar Lai. Indian J Microbiol 63, 33–41 (2023). https://doi.org/10.1007/s12088-022-01050-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-022-01050-9