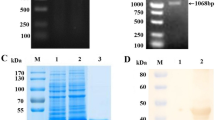
Abstract
In order to develop a more sensitive and reliable method for detection of serum antibodies against Mycoplasma hyopneumoniae infection in pigs, six recombinant proteins of M. hyopneumoniae (P102, P95, P46, P97 like, Lppt, and hypothetical P987) were used for the standardization of an indirect enzyme-linked immunosorbent assay (ELISA). The proteins were evaluated against 50 sera of the specific pathogen-free and 50 sera of pigs with lesions suggestive of infection. The sensitivity was 88%, 86%, 78%, 74%, 66%, and 60% for the proteins P102, P95, P46, P97 like, Lppt, and hypothetical protein P987, respectively. Moreover, the proteins were used to establish the seroprevalence in two different commercial herds (254 sera pigs from farm considered free of M. hyopneumoniae and 246 from farm with clinical signs of enzootic pneumonia and positive serology for M. hyopneumoniae) and the positive rate was 65.2% for P95, 54.6% for P102, 40.2% for P46, 37.2% for P97 like, 17.4% for the hypothetical P987, and 14% for Lppt protein. In addition, the ELISA with six recombinant proteins was compared to commercial HerdCheck kit using 118 random pig sera samples and the results showed that ELISA with recombinant proteins were more sensitive than the commercial test. These data show that the recombinant proteins P95 and P102 are potential targets to be used in diagnostic tests to detect antibodies against M. hyopneumoniae. Although more studies are necessary, this study provides insights that these recombinant proteins can be useful in epidemiological investigations and as potential biomarkers in differentiating infected animals from those vaccinated.



Similar content being viewed by others
Availability of data and material
Not Applicable.
References
Pieters MG, Maes D (2019) Mycoplasmosis. Dis Swine. https://doi.org/10.1002/9781119350927.ch56
Pieters M, Daniels J, Rovira A (2017) Comparison of sample types and diagnostic methods for in vivo detection of Mycoplasma hyopneumoniae during early stages of infection. Vet Microbiol 203:103–109. https://doi.org/10.1016/j.vetmic.2017.02.014
Poeta Silva APS, Magtoto RL, Souza Almeida HM et al (2020) Performance of commercial Mycoplasma hyopneumoniae serum enzyme-linked immunosorbent assays under experimental and field conditions. J Clin Microbiol 58:e00485–e00420. https://doi.org/10.1128/JCM.00485-20
Neto JCG, Strait EL, Raymond M et al (2014) Antibody responses of swine following infection with Mycoplasma hyopneumoniae, M. hyorhinis, M. hyosynoviae and M. flocculare. Vet Microbiol 174:163–171. https://doi.org/10.1016/j.vetmic.2014.08.008
Maes D, Sibila M, Kuhnert P et al (2018) Update on Mycoplasma hyopneumoniae infections in pigs: Knowledge gaps for improved disease control. Transbound Emerg Dis 65:110–124. https://doi.org/10.1111/tbed.12677
Sibila M, Pieters M, Molitor T et al (2009) Current perspectives on the diagnosis and epidemiology of Mycoplasma hyopneumoniae infection. Vet J 181:221–231. https://doi.org/10.1016/j.tvjl.2008.02.020
Liu W, Feng Z, Fang L et al (2011) Complete genome sequence of Mycoplasma hyopneumoniae strain 168. J Bacteriol 193:1016–1017. https://doi.org/10.1128/JB.01305-10
Minion FC, Lefkowitz EJ, Madsen ML et al (2004) The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol 186:7123–7133. https://doi.org/10.1128/JB.186.21.7123-7133.2004
Vasconcelos ATR, Ferreira HB, Bizarro CV et al (2005) Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J Bacteriol 187:5568–5577. https://doi.org/10.1128/JB.187.16.5568-5577.2005
Feng Z-X, Bai Y, Yao J-T et al (2014) Use of serological and mucosal immune responses to Mycoplasma hyopneumoniae antigens P97R1, P46 and P36 in the diagnosis of infection. Vet J 202:128–133. https://doi.org/10.1016/j.tvjl.2014.06.019
Simionatto S, Marchioro SB, Galli V et al (2012) Immunological characterization of Mycoplasma hyopneumoniae recombinant proteins. Comp Immunol Microbiol Infect Dis 35:209–216. https://doi.org/10.1016/j.cimid.2012.01.007
Simionatto S, Marchioro SB, Galli V et al (2010) Cloning and purification of recombinant proteins of Mycoplasma hyopneumoniae expressed in Escherichia coli. Protein Expr Purif 69:132–136. https://doi.org/10.1016/j.pep.2009.09.001
Wei C, Huang Z, Sun L et al (2013) Expression and antibody preparation of GP5a gene of porcine reproductive and respiratory syndrome virus. Indian J Microbiol 53:370–375. https://doi.org/10.1007/s12088-013-0368-1
Bantis LE, Nakas CT, Reiser B (2014) Construction of confidence regions in the ROC space after the estimation of the optimal Youden index-based cut‐off point. Biometrics 70:212–223. https://doi.org/10.1111/biom.12107
Tseng Y-H, Hsieh C-C, Kuo T-Y et al (2019) Construction of a Lactobacillus plantarum Strain expressing the capsid protein of porcine circovirus type 2d (PCV2d) as an oral vaccine. Indian J Microbiol 59:490–499. https://doi.org/10.1007/s12088-019-00827-9
Petersen AC, Oneal DC, Seibel JR et al (2016) Cross reactivity among the swine mycoplasmas as identified by protein microarray. Vet Microbiol 192:204–212. https://doi.org/10.1016/j.vetmic.2016.07.023
Meens J, Selke M, Gerlach G-F (2006) Identification and immunological characterization of conserved Mycoplasma hyopneumoniae lipoproteins Mhp378 and Mhp651. Vet Microbiol 116:85–95. https://doi.org/10.1016/j.vetmic.2006.03.011
Fisch A, Marchioro SB, Gomes CK et al (2016) Commercial bacterins did not induce detectable levels of antibodies in mice against Mycoplasma hyopneumoniae antigens strongly recognized by swine immune system. Trials Vaccinol 5:32–37. https://doi.org/10.1016/j.vetmic.2006.03.011
Ding H, Wen Y, Xu Z et al (2021) Development of an ELISA for distinguishing convalescent sera with Mycoplasma hyopneumoniae infection from hyperimmune sera responses to bacterin vaccination in pigs. Vet Med Sci. https://doi.org/10.1002/vms3.539
Feng Z-X, Shao G-Q, Liu M-J et al (2010) Development and validation of a SIgA-ELISA for the detection of Mycoplasma hyopneumoniae infection. Vet Microbiol 143:410–416. https://doi.org/10.1016/j.vetmic.2009.11.038
Bai Y, Gan Y, Hua L-Z et al (2018) Application of a sIgA-ELISA method for differentiation of Mycoplasma hyopneumoniae infected from vaccinated pigs. Vet Microbiol 223:86–92. https://doi.org/10.1016/j.vetmic.2018.07.023
Simionatto S, Marchioro SB, Maes D, Dellagostin OA (2013) Mycoplasma hyopneumoniae: From disease to vaccine development. Vet Microbiol 165:234–242. https://doi.org/10.1016/j.vetmic.2013.04.019
Bogema DR, Deutscher AT, Woolley LK et al Characterization of cleavage events in the multifunctional cilium adhesin Mhp684 (P146) reveals a rechanism by which Mycoplasma hyopneumoniae regulates surface topography. mBio 3:e00282-11. https://doi.org/10.1128/mBio.00282-11
Acknowledgements
We are grateful to Davi Barcellos and Andrea Panzardi for providing convalescent pig sera and Michele Ribeiro dos Santos for technical assistance.
Funding
This study was supported by the Brazilian National Research Council (CNPq grants 578614/2008–1 and 303148/2009–8), Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT grants 092/2015), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Contributions
Conceptualization, OAD, SBM, and SS; Methodology, CBB, MSB, SJ, and VG; Formal analysis and investigation, CBB, MSB, OAD, SBM, SJ, SS, and VG; Writing - original draft preparation, CBB, MSB, OAD, SBM, SJ, SS, and VG; Writing -review and editing, SBM, SS, and OAD; Funding acquisition, SBM, and SS; Supervision, SBM, and SS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None of the authors has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Ethics Approval
All animal procedures were performed at the animal facility of the Federal University of Pelotas (UFPel) and approved by the Ethics Committee for Animal Experimentation (CEEA) of UFPel under protocol number 5614. The CEEA of UFPel is accredited by the Brazilian National Council for Animal Experimentation Control (CONCEA).
Consent to Participate
Not Applicable.
Consent for Publication
Not Applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Simionatto, S., Marchioro, S.B., dos Santos Barbosa, M. et al. Development of ELISA Using Recombinant Proteins for the Diagnosis of Mycoplasma hyopneumoniae Infection. Indian J Microbiol 62, 88–95 (2022). https://doi.org/10.1007/s12088-021-00981-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-021-00981-z