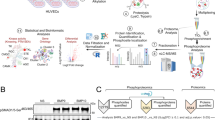
Abstract
Vascular endothelial growth factor-A (VEGF-A) is one of the primary factors promoting angiogenesis in endothelial cells. Although defects in VEGF-A signaling are linked to diverse pathophysiological conditions, the early phosphorylation-dependent signaling events pertinent to VEGF-A signaling remain poorly defined. Hence, a temporal quantitative phosphoproteomic analysis was performed in human umbilical vein endothelial cells (HUVECs) treated with VEGF-A-165 for 1, 5 and 10 min. This led to the identification and quantification of 1971 unique phosphopeptides corresponding to 961 phosphoproteins and 2771 phosphorylation sites in total. Specifically, 69, 153, and 133 phosphopeptides corresponding to 62, 125, and 110 phosphoproteins respectively, were temporally phosphorylated at 1, 5, and 10 min upon addition of VEGF-A. These phosphopeptides included 14 kinases, among others. This study also captured the phosphosignaling events directed through RAC, FAK, PI3K-AKT-MTOR, ERK, and P38 MAPK modules with reference to our previously assembled VEGF-A/VEGFR2 signaling pathway map in HUVECs. Apart from a significant enrichment of biological processes such as cytoskeleton organization and actin filament binding, our results also suggest a role of AAK1-AP2M1 in the regulation of VEGFR endocytosis. Taken together, the temporal quantitative phosphoproteomics analysis of VEGF signaling in HUVECs revealed early signaling events and we believe that this analysis will serve as a starting point for the analysis of differential signaling across VEGF members toward the full elucidation of their role in the angiogenesis processes.
Graphical abstract

Workflow for the identification of early phosphorylation events induced by VEGF-A-165 in HUVEC cells




Similar content being viewed by others
Data availability
The data supporting this study's finding is in this article and its supplementary information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD038133.
References
Abhinand CS, Raju R, Soumya SJ, Arya PS, Sudhakaran PR (2016) VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal 10:347–354. https://doi.org/10.1007/s12079-016-0352-8
Agajanian MJ, Walker MP, Axtman AD, Ruela-de-Sousa RR, Serafin DS, Rabinowitz AD, Graham DM, Ryan MB, Tamir T, Nakamichi Y, Gammons MV, Bennett JM, Couñago RM, Drewry DH, Elkins JM, Gileadi C, Gileadi O, Godoi PH, Kapadia N, Müller S, Santiago AS, Sorrell FJ, Wells CI, Fedorov O, Willson TM, Zuercher WJ, Major MB (2019) WNT activates the AAK1 kinase to promote clathrin-mediated endocytosis of LRP6 and establish a negative feedback loop. Cell Rep 26(1):79-93.e8. https://doi.org/10.1016/j.celrep.2018.12.023
Aguilar-Cazares D, Chavez-Dominguez R, Carlos-Reyes A, Lopez-Camarillo C, Hernadez de la Cruz ON, Lopez-Gonzalez JS (2019) Contribution of angiogenesis to inflammation and cancer. Front Oncol 12(9):1399. https://doi.org/10.3389/fonc.2019.01399
Bouïs D, Kusumanto Y, Meijer C, Mulder NH, Hospers GAP (2006) A review on pro- and anti-angiogenic factors as targets of clinical intervention. Pharmacol Res 53:89–103. https://doi.org/10.1016/j.phrs.2005.10.006
Bruce JI, Shuttleworth TJ, Giovannucci DR, Yule DI (2002) Phosphorylation of inositol 1,4,5-trisphosphate receptors in parotid acinar cells. A mechanism for the synergistic effects of cAMP on Ca2+ signaling. J Biol Chem 277(2):1340–1348. https://doi.org/10.1074/jbc.M106609200
Carmeliet P, Jain RK (2000) Angiogenesis in cancer and other diseases. Nature 407:249–257. https://doi.org/10.1038/35025220
Carmeliet P, Jain RK (2011) Molecular mechanisms and clinical applications of angiogenesis. Nature 473:298–307. https://doi.org/10.1038/nature10144
Chaar Z, O’reilly P, Gelman I, Sabourin LA (2006) v-Src-dependent down-regulation of the Ste20-like kinase SLK by casein kinase II. J Biol Chem 281(38):28193–28199. https://doi.org/10.1074/jbc.M605665200
Chatterjee S, Heukamp LC, Siobal M, Schöttle J, Wieczorek C, Peifer M, Frasca D, Koker M, König K, Meder L, Rauh D, Buettner R, Wolf J, Brekken RA, Neumaier B, Christofori G, Thomas RK, Ullrich RT (2013) Tumor VEGF: VEGFR2 autocrine feed-forward loop triggers angiogenesis in lung cancer. J Clin Invest 123:1732–1740. https://doi.org/10.1172/JCI65385
Cheng A, Grant CE, Noble WS, Bailey TL (2019) MoMo: discovery of statistically significant post-translational modification motifs. Bioinformatics 35(16):2774–2782. https://doi.org/10.1093/bioinformatics/bty1058
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Chou MF, Schwartz D (2011) Biological sequence motif discovery using motif-x. Curr Protoc Bioinf 13:15–24. https://doi.org/10.1002/0471250953.bi1315s35
Clarke DJB, Kuleshov MV, Schilder BM, Torre D, Duffy ME, Keenan AB, Lachmann A, Feldmann AS, Gundersen GW, Silverstein MC, Wang Z, Ma’ayan A (2018) eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res 46(W1):W171–W179. https://doi.org/10.1093/nar/gky458
Conner SD, Schmid SL (2002) Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol 156(5):921–929. https://doi.org/10.1083/jcb.200108123
Davis S, Aldrich TH, Jones PF, Acheson A, Compton DL, Jain V, Ryan TE, Bruno J, Radziejewski C, Maisonpierre PC, Yancopoulos GD (1996) Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell 87(7):1161–1169. https://doi.org/10.1016/s0092-8674(00)81812-7
Dejana E (2010) The role of wnt signaling in physiological and pathological angiogenesis. Circ Res 107:943–952. https://doi.org/10.1161/CIRCRESAHA.110.223750
Eid S, Turk S, Volkamer A, Rippmann F, Fulle S (2017) KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinf 18(1):16. https://doi.org/10.1186/s12859-016-1433-7
Eng JK, McCormack AL, Yates JR (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5(11):976–989. https://doi.org/10.1016/1044-0305(94)80016-2
Ferrara N (1999) Molecular and biological properties of vascular endothelial growth factor. J Mol Med 77:527–543. https://doi.org/10.1007/s001099900019
Ferrara N, Houck K, Jakeman L, Leung DW (1992) Molecular and biological properties of the vascular endothelial growth factor family of proteins. Endocr Rev 13:18–32. https://doi.org/10.1210/edrv-13-1-18
Folkman J (1971) Tumor angiogenesis: therapeutic implications. N Engl J Med 285:1182–1186. https://doi.org/10.1056/NEJM197111182852108
Folkman J, Klagsburn M (1987) Angiogenic factors. Science 235(4787):442–447. https://doi.org/10.1126/science.2432664
Folkman J, Shing Y (1992) Angiogenesis. J Biol Chem 267(16):10931–10934. https://doi.org/10.1016/S0021-9258(19)49853-0
Grønborg M, Kristiansen TZ, Stensballe A, Andersen JS, Ohara O, Mann M, Jensen ON, Pandey A (2002) A mass spectrometry-based proteomic approach for identification of serine/threonine-phosphorylated proteins by enrichment with phospho-specific antibodies: identification of a novel protein, Frigg, as a protein kinase A substrate. Mol Cell Proteomics 1(7):517–527. https://doi.org/10.1074/mcp.m200010-mcp200
Holmes K, Roberts OL, Thomas AM, Cross MJ (2007) Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 19:2003–2012. https://doi.org/10.1016/j.cellsig.2007.05.013
Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L (2011) Signal transduction by vascular endothelial growth factor receptors. Biochem J 437:169–183. https://doi.org/10.1042/BJ20110301
Koenig T, Menze BH, Kirchner M, Monigatti F, Parker KC, Patterson T, Steen JJ, Hamprecht FA, Steen H (2008) Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J Proteome Res 7(9):3708–3717. https://doi.org/10.1021/pr700859x
Lohela M, Bry M, Tammela T, Alitalo K (2009) VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol 21:154–165. https://doi.org/10.1089/15279160175029434410.1016/j.ceb.2008.12.012
Mann M, Jensen ON (2003) Proteomic analysis of post-translational modifications. Nat Biotechnol 21(3):255–261. https://doi.org/10.1038/nbt0303-255
Ng EW, Adamis AP (2005) Targeting angiogenesis, the underlying disorder in neovascular age-related macular degeneration. Can J Ophthalmol 40(3):352–368. https://doi.org/10.1016/S0008-4182(05)80078-X
Nishida N, Yano H, Nishida T, Kamura T, Kojiro M (2006) Angiogenesis in cancer. Vasc Health Risk Manag 2:213–219. https://doi.org/10.2147/vhrm.2006.2.3.213
Nugent MA, Iozzo RV (2000) Fibroblast growth factor-2. Int J Biochem Cell Biol 32(2):115–120. https://doi.org/10.1016/s1357-2725(99)00123-5
O’Flaherty C, de Lamirande E, Gagnon C (2004) Phosphorylation of the Arginine-X-X-(Serine/Threonine) motif in human sperm proteins during capacitation: modulation and protein kinase a dependency. Mol Hum Reprod 10(5):355–363. https://doi.org/10.1093/molehr/gah046
Oklu R, Walker TG, Wicky S, Hesketh R (2010) Angiogenesis and current antiangiogenic strategies for the treatment of cancer. J Vasc Interv Radiol 21(12):1791–1805. https://doi.org/10.1016/j.jvir.2010.08.009
Ortega N, Hutchings H, Plouët J (1999) Signal relays in the VEGF system. Front Biosci. 1(4):D141–D152. https://doi.org/10.2741/A417
Patil AH, Datta KK, Behera SK, Kasaragod S, Pinto SM, Koyangana SG, Mathur PP, Gowda H, Pandey A, Prasad TSK (2018) Dissecting Candida pathobiology: post-translational modifications on the candida tropicalis proteome. OMICS 22(8):544–552. https://doi.org/10.1089/omi.2018.0093
Pawlowska Z, Baranska P, Jerczynska H, Koziolkiewicz W, Cierniewski CS (2005) Heat shock proteins and other components of cellular machinery for protein synthesis are up-regulated in vascular endothelial cell growth factor-activated human endothelial cells. Proteomics 5(5):1217–1227. https://doi.org/10.1002/pmic.200400983
Petrova TV, Makinen T, Alitalo K (1999) Signaling via vascular endothelial growth factor receptors. Exp Cell Res 253(1):117–130. https://doi.org/10.1006/excr.1999.4707
Rahimi N, Costello CE (2015) Emerging roles of post-translational modifications in signal transduction and angiogenesis. Proteomics 15(2–3):300–309. https://doi.org/10.1002/pmic.201400183
Ramsbottom KA, Prakash A, Riverol YP, Camacho OM, Martin MJ, Vizcaíno JA, Deutsch EW, Jones AR (2022) Method for independent estimation of the false localization rate for phosphoproteomics. J Proteome Res 21(7):1603–1615. https://doi.org/10.1021/acs.jproteome.1c00827
Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75(3):663–670. https://doi.org/10.1021/ac026117i
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists. Nucleic Acids Res 47(W1):W191–W198. https://doi.org/10.1093/nar/gkz369
Santarpia L, Lippman SM, El-Naggar AK (2012) Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 16(1):103–119. https://doi.org/10.1517/14728222.2011.645805
Seldin DC, Leder P (1994) Mutational analysis of a critical signaling domain of the human interleukin 4 receptor. Proc Natl Acad Sci U S A 91(6):2140–2144. https://doi.org/10.1073/pnas.91.6.2140
Soumya SJ, Athira AP, Binu S, Sudhakaran PR (2016) mTOR as a modulator of metabolite sensing relevant to angiogenesis. In: Maiese K (ed) Molecules to medicine with mTOR: Translating critical pathways of the mammalian target of rapamycin into novel therapeutic strategies. Elsevier Science & Technology Books. Academic Press, pp 229–243
Sparks LM, Moro C, Ukropcova B, Bajpeyi S, Civitarese AE, Hulver MW, Thoresen GH, Rustan AC, Smith SR (2011) Remodeling lipid metabolism and improving insulin responsiveness in human primary myotubes. PLoS ONE 6(7):e21068. https://doi.org/10.1371/journal.pone.0021068
Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics 6(6):1103–1109. https://doi.org/10.1074/mcp.T600060-MCP200
Sunitha P, Raju R, Sajil CK, Abhinand CS, Nair AS, Oommen OV, Sugunan VS, Sudhakaran PR (2019) Temporal VEGFA responsive genes in HUVECs: gene signatures and potential ligands/receptors fine-tuning angiogenesis. J Cell Commun Signal 13(4):561–571. https://doi.org/10.1007/s12079-019-00541-7
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612. https://doi.org/10.1093/nar/gkaa1074
Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10(12):5354–5362. https://doi.org/10.1021/pr200611n
Tonnesen MG, Feng X, Clark RAF (2000) Angiogenesis in wound healing. J Investig Dermatol Symp Proc 5:40–46. https://doi.org/10.1046/j.1087-0024.2000.00014.x
Torres M (2003) Mitogen-activated protein kinase pathways in redox signaling. Front Biosci 8:d369–d391. https://doi.org/10.2741/999
Wagner S, Storbeck CJ, Roovers K, Chaar ZY, Kolodziej P, McKay M, Sabourin LA (2008) FAK/src-family dependent activation of the Ste20-like kinase SLK is required for microtubule-dependent focal adhesion turnover and cell migration. PLoS ONE 3(4):e1868. https://doi.org/10.1371/journal.pone.0001868
Wang Y, Singh AR, Zhao Y, Du T, Huang Y, Wan X, Mukhopadhyay D, Wang Y, Wang N, Zhang P (2020) TRIM28 regulates sprouting angiogenesis through VEGFR-DLL4-Notch signaling circuit. FASEB J 34(11):14710–14724. https://doi.org/10.1096/fj.202000186RRR
Wu CF, Wang R, Liang Q, Liang J, Li W, Jung SY, Qin J, Lin SH, Kuang J (2010) Dissecting the M phase-specific phosphorylation of serine-proline or threonine-proline motifs. Mol Biol Cell 21(9):1470–1481. https://doi.org/10.1091/mbc.e09-06-0486
Yu S, Oh J, Li F, Kwon Y, Cho H, Shin J, Lee SK, Kim S (2017) New scaffold for angiogenesis inhibitors discovered by targeted chemical transformations of wondonin natural products. ACS Med Chem Lett 8(10):1066–1071. https://doi.org/10.1021/acsmedchemlett.7b00281
Zhuang G, Yu K, Jiang Z, Chung A, Yao J, Ha C, Toy K, Soriano R, Haley B, Blackwood E, Sampath D, Bais C, Lill JR, Ferrara N (2013) Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci Signal 6(271):ra25. https://doi.org/10.1126/scisignal.2003572
Acknowledgements
The authors would like to thank Ms. Sumiko Ohnuma for her support in preparing samples. This work was supported, in part, by research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan, and by a runner-up award fund from Kuraray/Leave a Nest to Chandran S. Abhinand in 2017. The authors also thank Karnataka Biotechnology and Information Technology Services (KBITS) and the Government of Karnataka for their support of the Center for Systems Biology and Molecular Medicine at Yenepoya (Deemed to be University) under the Biotechnology Skill Enhancement Program in Multiomics Technology (BiSEP GO ITD 02 MDA 2017).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
This article does not contain any studies with human participants or animals performed by any authors. The authors have no financial or proprietary interests in any of the material discussed in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abhinand, C.S., Galipon, J., Mori, M. et al. Temporal phosphoproteomic analysis of VEGF-A signaling in HUVECs: an insight into early signaling events associated with angiogenesis. J. Cell Commun. Signal. 17, 1067–1079 (2023). https://doi.org/10.1007/s12079-023-00736-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-023-00736-z