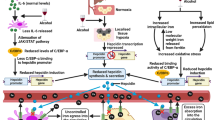
Abstract
Accumulating evidence shows that the intestinal microbiota is closely related to the pathophysiology and the disease progression of chronic hepatitis B virus (HBV) infection. The intestinal microbiota acts on the host through its metabolites. This review aimed to discuss the effects of gut microbiota metabolites on the disease progression of chronic HBV infection. A literature search on PubMed database and Wiley Online Library with pre-specified criteria yielded 96 unique results. After consensus by all authors, the contents from 86 original publications were extracted and included in this review. In liver disease with HBV infection, the intestinal microbiota changed in different stages and affected the production of bacterial metabolites. The abundance of bacteria producing short-chain fatty acids such as butyrate reduced, which was associated with bacterial translocation and the progression of liver disease. The intestinal microbiota—bile acid—host axis was destroyed, affecting the progression of the disease. Under the control of intestinal microbiota, tryptophan affected the gut–liver axis through three main metabolic pathways, among which the kynurenine pathway was closely related to the immune response of hepatitis B. The level of trimethylamine-N-oxide decreased in liver cancer with HBV infection and were used as a potential biomarker of liver cancer. Vitamin deficiencies, including those of vitamin D and vitamin A related to microbiota, were common and associated with survival. Hydrogen sulfide regulated by the intestinal microbiota was also closely related to the gut–liver axis. In liver disease with hepatitis B infection, the intestinal microbiota is imbalanced, and a variety of intestinal microbiota metabolites participate in the occurrence and development of the disease.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- Ac-p53:
-
Acetylated p53
- AMPs:
-
Antimicrobial peptides
- AhR:
-
Aryl hydrocarbon Receptor
- AhR-IL22:
-
AhR-dependent interleukin 22
- BAs:
-
Bile acids
- BSH:
-
Bile salt hydrolase
- CHB:
-
Chronic hepatitis B
- Conv-R mice:
-
Conventionally raised mice
- CYP7A1:
-
Cholesterol 7α-hydroxylase
- CXCL:
-
Chemokine C-X-C motif ligand
- CSE:
-
Cystathionine γ-lyase
- DOX:
-
Doxycycline
- DAC:
-
Deoxycholic acid
- FXR:
-
Farnesol-X receptor
- FGF15:
-
Fibroblast growth factor 15
- FGF19:
-
Fibroblast growth factor 19
- GLP-1:
-
Glucagon-like peptide-1
- GF mice:
-
Germ-free mice
- HDAC:
-
Histone deacetylase
- 5-HT:
-
5-Hydroxytryptamine
- H2S:
-
Hydrogen sulfide
- IDO-1:
-
Indoleamine 2,3-dioxygenase-1
- Kyn:
-
Kynurenine
- KP:
-
Kynurenine pathway
- Kyn/Try:
-
Kynurenine/tryptophan
- LCMV:
-
Lymphocytic choriomeningitis virus
- mTOR:
-
Mammalian target of rapamycin
- NTCP:
-
Sodium taurocholate cotransport peptides
- PCLS:
-
Precision-cut liver slices
- peg-IFN:
-
Pegylated interferon-α 2a
- SCFAs:
-
Short-chain fatty acids
- SPF mice:
-
Specific pathogen-free mice
- SIRT-1:
-
Sirtuin-1
- SHP:
-
Small heterodimer partner
- SSRIs:
-
Selective serotonin reuptake inhibitors
- TGR5:
-
Takeda G-protein coupled receptor 5
- Trp:
-
Tryptophan
- TUDCA:
-
Tauroursodeoxycholic acid
- TDO:
-
Tryptophan 2,3-dioxygenase
- TNF-a:
-
Tumor necrosis factor a
- TPH1:
-
Tryptophan hydroxylase 1
- TMA:
-
Trimethylamine
- TMAO:
-
Trimethylamine-N-oxide
References
Zhu Q, Xia P, Zhou X, et al. Hepatitis B virus infection alters gut microbiota composition in mice. Front Cell Infect Microbiol 2019;9:377
Yang XA, Lv F, Wang R, et al. Potential role of intestinal microflora in disease progression among patients with different stages of Hepatitis B. Gut Pathog 2020;12:50
Chen Z, Xie Y, Zhou F, et al. Featured gut microbiomes associated with the progression of chronic hepatitis B disease. Front Microbiol 2020;11:383
Heintz-Buschart A, Wilmes P. Human gut microbiome: function matters. Trends Microbiol 2018;26(7):563–574
Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol 2017;35:8–15
Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16(6):341–352
Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336(6086):1262–1267
Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol 2011;61(3):693–703
Mohamadkhani A. On the potential role of intestinal microbial community in hepatocarcinogenesis in chronic hepatitis B. Cancer Med 2018. https://doi.org/10.1002/cam4.1550
Zeng Y, Chen S, Fu Y, et al. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J Viral Hepat 2020;27(2):143–155
Donia MS, Fischbach MA. Small molecules from the human microbiota. Science 2015;349(6246):1254766
Kaitlyn O, Emma A. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 2019. https://doi.org/10.1186/s40168-019-0704-8
Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165(6):1332–1345
Patrice D, Cani MH, Charlotte Lefort C, Marialetizia RA. Microbial regulation of organismal energy. Nat Metab 2019;1:34–46
Jung TH, Park JH, Jeon WM, Han KS. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract 2015;9(4):343–349
Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 2009;139(9):1619–1625
Schulthess J, Pandey S, Capitani M, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 2019;50(2):432-445.e7
Martin-Gallausiaux C, Larraufie P, Jarry A, et al. Butyrate produced by commensal bacteria down-regulates indolamine 2,3-dioxygenase 1 (IDO-1) expression via a dual mechanism in human intestinal epithelial cells. Front Immunol 2018;9:2838
Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019;68(6):1014–1023
Pant K, Yadav AK, Gupta P, Islam R, Saraya A, Venugopal SK. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol 2017;12:340–349
Liu H, Wang J, He T, et al. Butyrate: a double-edged sword for health. Adv Nutr 2018;9(1):21–29
Pant K, Mishra AK, Pradhan SM, et al. Butyrate inhibits HBV replication and HBV-induced hepatoma cell proliferation via modulating SIRT-1/Ac-p53 regulatory axis. Mol Carcinog 2019;58(4):524–532
Jenessa A, Winston CMT. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2019. https://doi.org/10.1080/19490976.2019.1674124
Ridlon Jason M, Harris Spencer C, Shiva B, Dae-Joong K, Hylemon Phillip B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016. https://doi.org/10.1080/19490976.2015.1127483
Makoto U, Makoto M, Hayato Y. Gut microbiota and host metabolism in liver cirrhosis. World J Gastroenterol 2015;21(41):11597–11608
Long Sarah L, Gahan Cormac GM, Joyce Susan A. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med 2017. https://doi.org/10.1016/j.mam.2017.06.002
Mullish Benjamin H, Alexandros P, Barker Grace F, Thursz Mark R, Marchesi Julian R, McDonald Julie AK. Functional microbiomics: evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods 2018. https://doi.org/10.1016/j.ymeth.2018.04.028
Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 2013;17(2):225–235
Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 2006;103(10):3920–3925
Huang HJ, Zhang AY, Cao HC, et al. Metabolomic analyses of faeces reveals malabsorption in cirrhotic patients. Dig Liver Dis 2013;45(8):677–682
Wang X, Chen L, Wang H, Cai W, Xie Q. Modulation of bile acid profile by gut microbiota in chronic hepatitis B. J Cell Mol Med 2020;24(4):2573–2581
Lorenzo-Zúñiga V, Bartolí R, Planas R, et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology 2003;37(3):551–557
Wang X, Xie G, Zhao A, et al. Serum bile acids are associated with pathological progression of hepatitis B-induced cirrhosis. J Proteome Res 2016;15(4):1126–1134
Petrick JL, Florio AA, Koshiol J, et al. Prediagnostic concentrations of circulating bile acids and hepatocellular carcinoma risk: REVEAL-HBV and HCV studies. Int J Cancer 2020;147(10):2743–2753
Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013;499(7456):97–101
Mouzannar K, Fusil F, Lacombe B, et al. Farnesoid X receptor-α is a proviral host factor for hepatitis B virus that is inhibited by ligands in vitro and in vivo. FASEB J 2019;33(2):2472–2483
Yan H, Zhong G, Xu G, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. https://doi.org/10.7554/eLife.00049
König A, Döring B, Mohr C, Geipel A, Geyer J, Glebe D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol 2014;61(4):867–875
Yan H, Peng B, Liu Y, et al. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J Virol 2014;88(6):3273–3284
Oehler N, Volz T, Bhadra OD, et al. Binding of hepatitis B virus to its cellular receptor alters the expression profile of genes of bile acid metabolism. Hepatology 2014;60(5):1483–1493
Radreau P, Porcherot M, Ramière C, Mouzannar K, Lotteau V, André P. Reciprocal regulation of farnesoid X receptor α activity and hepatitis B virus replication in differentiated HepaRG cells and primary human hepatocytes. FASEB J 2016;30(9):3146–3154
Robin E PA, Elise R NB, Hugo G RD, Jacky V. Safety and antiviral effect of the farnesoid X receptor agonist EYP001 in chronic hepatitis B patients: a randomised placebo-controlled phase 1b study. Ameican Association for the study of Liver Diseases Abstracts 709, 2019.
Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun 2018;9(1):3294
Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018;23(6):716–724
Mao R, Zhang J, Jiang D, et al. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J Virol 2011;85(2):1048–1057
Yoshio S, Sugiyama M, Shoji H, et al. Indoleamine-2,3-dioxygenase as an effector and an indicator of protective immune responses in patients with acute hepatitis B. Hepatology 2016;63(1):83–94
Mehraj V, Routy JP. Tryptophan catabolism in chronic viral infections: handling uninvited guests. Int J Tryptophan Res 2015;8:41–48
Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013;39(2):372–385
Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22(6):598–605
Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA 2010;107(1):228–233
Shimada Y, Kinoshita M, Harada K, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 2013;8(11):e80604
Beaumont M, Neyrinck AM, Olivares M, et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J 2018;32:6681–6693
Huc T, Konop M, Onyszkiewicz M, et al. Colonic indole, gut bacteria metabolite of tryptophan, increases portal blood pressure in rats. Am J Physiol Regul Integr Comp Physiol 2018;315(4):R646–R655
Gao J, Xu K, Liu H, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol 2018;8:13
Badawy AA. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int J Tryptophan Res 2017;10:1178646917691938
Van der Leek AP, Yanishevsky Y, Kozyrskyj AL. The kynurenine pathway as a novel link between allergy and the gut microbiome. Front Immunol 2017;8:1374
Clarke G, Grenham S, Scully P, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2013;18(6):666–673
Desbonnet L, Clarke G, Traplin A, et al. Gut microbiota depletion from early adolescence in mice: implications for brain and behaviour. Brain Behav Immun 2015;48:165–173
Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 2017;112(Pt B):399–412
Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015;161(2):264–276
Kwon YH, Wang H, Denou E, et al. Modulation of gut microbiota composition by serotonin signaling influences intestinal immune response and susceptibility to colitis. Cell Mol Gastroenterol Hepatol 2019;7(4):709–728
Lesurtel M, Soll C, Humar B, Clavien PA. Serotonin: a double-edged sword for the liver. Surgeon 2012;10(2):107–113
Lang PA, Contaldo C, Georgiev P, et al. Aggravation of viral hepatitis by platelet-derived serotonin. Nat Med 2008;14(7):756–761
Chang CM, Hsieh MS, Yang TC, et al. Selective serotonin reuptake inhibitors and the risk of hepatocellular carcinoma in hepatitis B virus-infected patients. Cancer Manag Res 2017;9:709–720
Canyelles M, Tondo M, Cedó L, Farràs M, Escolà-Gil JC, Blanco-Vaca F. Trimethylamine N-Oxide: a link among diet, gut microbiota, gene regulation of liver and intestine cholesterol homeostasis and HDL function. Int J Mol Sci 2018. https://doi.org/10.3390/ijms19103228
Chittim CL, Martínez Del Campo A, Balskus EP. Gut bacterial phospholipase Ds support disease-associated metabolism by generating choline. Nat Microbiol 2019;4(1):155–163
Romano KA, Martinez-Del Campo A, Kasahara K, et al. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 2017;22(3):279-290.e7
Cox IJ, Aliev AE, Crossey MM, et al. Urinary nuclear magnetic resonance spectroscopy of a Bangladeshi cohort with hepatitis-B hepatocellular carcinoma: a biomarker corroboration study. World J Gastroenterol 2016;22(16):4191–4200
Steinert RE, Lee YK, Sybesma W. Vitamins for the gut microbiome. Trends Mol Med 2020;26(2):137–140
Cantorna MT, Snyder L, Arora J. Vitamin A and vitamin D regulate the microbial complexity, barrier function, and the mucosal immune responses to ensure intestinal homeostasis. Crit Rev Biochem Mol Biol 2019;54(2):184–192
Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 2015;6:148
Karl JP, Meydani M, Barnett JB, et al. Fecal concentrations of bacterially derived vitamin K forms are associated with gut microbiota composition but not plasma or fecal cytokine concentrations in healthy adults. Am J Clin Nutr 2017;106(4):1052–1061
Koop AH, Mousa OY, Pham LE, Corral-Hurtado JE, Pungpapong S, Keaveny AP. An argument for vitamin D, A, and zinc monitoring in cirrhosis. Ann Hepatol 2018;17(6):920–932
Hoan NX, Khuyen N, Binh MT, et al. Association of vitamin D deficiency with hepatitis B virus—related liver diseases. BMC Infect Dis 2016;16(1):507
Shen X, Carlström M, Borniquel S, Jädert C, Kevil CG, Lundberg JO. Microbial regulation of host hydrogen sulfide bioavailability and metabolism. Free Radic Biol Med 2013;60:195–200
Wallace JL, Motta JP, Buret AG. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am J Physiol Gastrointest Liver Physiol 2018;314(2):G143–G149
Kalantar-Zadeh K, Berean KJ, Burgell RE, Muir JG, Gibson PR. Intestinal gases: influence on gut disorders and the role of dietary manipulations. Nat Rev Gastroenterol Hepatol 2019;16(12):733–747
Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 2012;3:448
Blachier F, Beaumont M, Kim E. Cysteine-derived hydrogen sulfide and gut health: a matter of endogenous or bacterial origin. Curr Opin Clin Nutr Metab Care 2019;22(1):68–75
Wu DD, Wang DY, Li HM, Guo JC, Duan SF, Ji XY. Hydrogen sulfide as a novel regulatory factor in liver health and disease. Oxid Med Cell Longev 2019;2019:3831713
Fan HN, Wang HJ, Yang-Dan CR, et al. Protective effects of hydrogen sulfide on oxidative stress and fibrosis in hepatic stellate cells. Mol Med Rep 2013;7(1):247–253
Zhang F, Jin H, Wu L, et al. Diallyl trisulfide suppresses oxidative stress-induced activation of hepatic stellate cells through production of hydrogen sulfide. Oxid Med Cell Longev 2017. https://doi.org/10.1155/2017/1406726
Wei W, Wang C, Li D. The content of hydrogen sulfide in plasma of cirrhosis rats combined with portal hypertension and the correlation with indexes of liver function and liver fibrosis. Exp Ther Med 2017;14(5):5022–5026
Barton LL, Ritz NL, Fauque GD, Lin HC. Sulfur cycling and the intestinal microbiome. Dig Dis Sci 2017;62(9):2241–2257
Wang J, Wang Y, Zhang X, et al. Gut microbial dysbiosis is associated with altered hepatic functions and serum metabolites in chronic hepatitis B patients. Front Microbiol 2017;8:2222
Ren YD, Ye ZS, Yang LZ, et al. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology 2017;65(5):1765–1768
Funding
This work was supported by the National Science and Technology Major Project of China under Grant (2018ZX10302-206-003-006); the Capital's Funds for Health Improvement and Research under Grant (CFH 2020-1-2171 and CFH 2018-2-2173); the Beijing Hospitals Authority Clinical Medicine Development of Special Funding under Grant (XMLX201837); the Digestive Medical Coordinated Development Center of Beijing Hospitals Authority under Grant (XXT26).
Author information
Authors and Affiliations
Contributions
HX, CQP and XS had designed the article, XS performed the literature search and data analysis, and XS drafted the manuscript and HX, CQP critically revised the manuscript for important intellectual content and finally approved the version to be submitted.
Corresponding authors
Ethics declarations
Conflict of interest
Xiu Sun, Cavin.Q.Pan and Huichun Xing declare that there is no conflict of interest regarding the publication of this article.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors approved the version of the manuscript to be published.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, X., Pan, C.Q. & Xing, H. Effect of microbiota metabolites on the progression of chronic hepatitis B virus infection. Hepatol Int 15, 1053–1067 (2021). https://doi.org/10.1007/s12072-021-10230-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-021-10230-6