Abstract
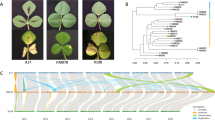
Powdery mildew (PM) is one of the most destructive diseases affecting rubber trees (Hevea brasiliensis), leading to severe yield losses. Sulfur dusting is used to control the propagation of the disease, and no specific fungicides have been developed yet. Therefore, identification of genetic level disease resistance in the rubber trees is very crucial. In this study, we describe the whole-genome sequencing of a PM resistant rubber clone, RRIC 52, as well as a PM susceptible clone, PB 235, using massively parallel paired-end sequencing. The MLO (mildew resistance locus O) genes were identified and analyzed for their structural features, and the variations were compared in the MLO gene family between the two clones. Sequencing results showed that RRIC 52 contained 4,280,477 SNPs and 400,667 InDels, while PB 235 contained 3,651,524 SNPs and 318,899 InDels. We identified 34 MLO genes (HbMLO 1 to 34) in the rubber genome. Multiple sequence alignment identified the conserved MLO domain and its TM domain in all HbMLO proteins. Sequence analysis identified non-synonymous variations (NSVs) in 12 HbMLO proteins. Phylogenetic analysis of the HBMLO genes revealed seven different clades. Six HbMLO genes in Clade V were orthologous to Arabidopsis genes where PM interaction was previously identified. The outcomes of this study widen the understanding of the MLO gene family, which can be used in breeding disease resistant rubber varieties in the future.




Similar content being viewed by others
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol 204(2):273–281
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P (1997) The barley MLO gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Chen Y, Wang Y, Zhang H (2014) Genome-wide analysis of the mildew resistance locus o (‘MLO’) gene family in tomato (‘Solanum lycopersicum’L.). Plant Omics 7(2):87
Consonni C, Bednarek P, Humphry M, Francocci F, Ferrari S, Harzen A, Panstruga R (2010) Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol 152(3):1544–1561
Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Panstruga R (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38(6):716–720. https://doi.org/10.1038/ng1806
Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, Goh CS, Cohen FE, Emerson BC, Schulze-Lefert P, Panstruga R (2003) Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol 56:77–88
Devoto A, Piffanelli P, Nilsson IM, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the MLO family in plants. J Biol Chem 274:34993–35004
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB (2008) Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol 35:1255–1266
Fernando TM (1971) Oidium leaf disease-the effect of environment and control measures on incidence of disease and atmospheric spore concentration. Quart J Rubb Res Inst Ceylon 48:100–111
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Bateman A (2016) The Pfam protein families’ database: towards a more sustainable future. Nucleic Acids Res 44(1):279–285. https://doi.org/10.1093/nar/gkv1344
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Jayasinghe CK (2001) Common disease. In: Tillekaratne LMK, Nugawela A (eds) Hand book of rubber agronomy, vol 1. Rubber Research Institute of Sri Lanka, Sri Lanka, pp 97–105
Jorgensen JH (1992) Discovery, characterization and exploitation of MLO powdery mildew resistance in barley. Euphytica 63:141–152
Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS, Moon BC, Kang CH, Park CY, Yoo JH, Kang YH, Koo SC, Koo YD, Jung JC, Kim ST, Schulze-Lefert P, Lee SY, Cho MJ (2002) MLO, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. J Biol Chem 277:19304–19314
Konishi S, Sasakuma T, Sasanuma T (2010) Identifcation of novel Mlo family members in wheat and their genetic characterization. Genes Genet Sys 85:167–175
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14):1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079
Liyanage AS, Kuruvilla JC (1992) Diseases of economic importance in rubber. In: Sethuraj MR, Mathew NM (eds) Natural rubber: biology, cultivation and technology, developments in crop science, vol 23. Elsevier, The Netherland, pp 324–359
Liyanage KK, Khan S, Mortimer PE, Hyde KD, Xu J, Brooks S, Ming Z (2016) Powdery mildew disease of rubber tree. Forest Pathol 46(2):90–103
Liyanage KK, Khan S, Brooks S, Mortimer PE, Karunarathna SC, Xu J, Hyde KD (2018) Morpho-molecular characterization of two Ampelomyces spp. (Pleosporales) strains. Mycoparasites of powdery mildew of Hevea brasiliensis. Front Microbiol 9:12. https://doi.org/10.3389/fmicb.2018.00012
Liyanage KK, Khan S, Brooks S, Mortimer PE, Karunarathna SC, Xu J, Hyde KD (2017) Taxonomic revision and phylogenetic analyses of rubber powdery mildew fungi. Microb Pathog 105:185–195
Min T, Haibin C, Aihua S, Huisun H (2012) In vitro culture method of powdery mildew (O. heveae Stanmann) of Hevea brasiliensis. Afr J Biotechnol 11(65):13167–13172
Mistry J, Finn RD, Eddy SR, Bateman A, Punta M (2013) Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41(12):e121. https://doi.org/10.1093/nar/gkt263
Moore AD, Heldy A, Terrapon N, Weiner J, Bornberg-Bauer E. (2014) DoMosaics: software for domain arrangement visualization and domain-centric analysis of proteins. Bioinformatics, 30(2): 282–283. https://doi.org/10.1093/bioinformatics/ btt640
Narayanan C, Mydin KK (2012) Breeding for disease resistance in Hevea spp.-status, potential threats, and possible strategies. In: Sniezko RA, Yanchuk AD, Kliejunas JT, Palmieri KM, Alexander JM, Frankel SJ, tech. coords (eds) Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: Disease and insect resistance in forest trees. Gen. Tech. Rep. PSW-GTR-240, 240. Pacific Southwest Research Station, Forest Service, US Department of Agriculture, Albany, CA, pp 240-251
Okonechnikov K, Golosova O, Fursov M, Varlamov A, Vaskin Y, Efremov I, Tleukenov T (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Omasits U, Ahrens CH, Müller S, Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30(6):884–886. https://doi.org/10.1093/bioinformatics/btt607
Panstruga R (2005) Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol 59(3):485–500
Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129(3):1076–1085
Qin B, Zheng F, Zhang Y (2015) Molecular cloning and characterization of a Mlo gene in rubber tree (Hevea brasiliensis). J Plant Physiol 175:78–85
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol 7(539):539. https://doi.org/10.1038/msb.2011.75
Silvestro D, Michalak I (2012) RaxmlGUI: a graphical front-end for RAxML. Org Diver Evol 12(4):335–337. https://doi.org/10.1007/s13127-011-0056-0
Tang C, Yang M, Fang Y, Luo Y et al (2016) The rubber tree genome reveals new insight into rubber production and species adaptation. Nat Plants 2:16073. https://doi.org/10.1038/NPLANTS.2016.73
Wastie RL (1975) Disease of rubber and their control. Tropical Pest Manag 21(3):268–288
Wolter M, Hollricher K, Salamini F, Schulze-Lefert P (1993) The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Genet Genomics 239:122–128
Acknowledgments
This work was supported by Key Research Program of Frontier Sciences, CAS, grant No. QYZDY-SSW-SMC014. We would like to thank the National Science Foundation of China (NSFC) for funding this work under the project codes Y4ZK111B01, 41761144055, 41771063, 31650410651, 41761144055 and 31550110215. Key Research Program of the Ministry of Sciences and Technology (Grant No. 2017YFC0505101) of China partially supported this work.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by: Jorge M. Santamaria
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liyanage, K.K., Khan, S., Herath, V. et al. Genome Wide Identification of the MLO Gene Family Associated with Powdery Mildew Resistance in Rubber Trees (Hevea brasiliensis). Tropical Plant Biol. 13, 331–342 (2020). https://doi.org/10.1007/s12042-020-09262-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-020-09262-3