Abstract
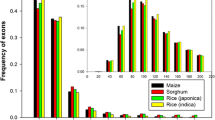
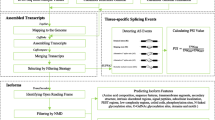
Pineapple (Ananas comosus L. Merrill) is an important tropical and subtropical fruit crop and possesses crassulacean acid metabolism (CAM) photosynthesis. Recent release of its genome sequences makes it possible to identify genes transcribed with alternatively spliced isoforms in this plant. Mapping the assembled transcripts generated by next-generation sequencing technology and existing expressed sequence tags as well as mRNA sequences to the published pineapple genome, we identified and analyzed alternative splicing (AS) events. We identified a total of 10,348 AS events involving 13,449 assembled putative unique transcripts, which were mapped to 5146 pineapple gene models that equivalent to 29.7 % of total expressed gene models. Consistent with previous findings in other plant species, intron retention (61.9 %) remains to be the dominant type among the identified AS events. Comparative genomic analysis of genes which generated pre-mRNAs having AS revealed a total of 481 genes conserved among Oryza sativa (ssp japonica), Sorghum bicolor, Zea mays, and pineapple, with 51 of them were also conserved with Brachypodium distachyon. Gene Ontology classification revealed that the products of these genes which generate AS isoforms are involved in many biological processes with diverse molecular functions. We annotated all assembled transcripts and also associated them with predicted gene models. The annotated information of these data provides a resource for further characterizing these genes and their biological roles. The data can be accessed at Plant Alternative Splicing Database (http://proteomics.ysu.edu/altsplice/).



Similar content being viewed by others
Abbreviations
- AS:
-
Alternative splicing
- ESTs:
-
Expressed sequence tags
- GO:
-
Gene ontology
- PUT:
-
Putative unique transcript
- FPKM:
-
Fragments Per Kilobase of exon model per Million mapped reads
- rpsBLAST:
-
Reversed position specific BLAST.
References
Bartholomew DP (2013) History and perspectives on the role of ethylene in pineapple flowering. In: XII international symposium on plant Bioregulators in fruit production. Acta Hortic 1042:269–284
Bartholomew DP, Kadzimin SB (1977) Pineapple. In: Alvin PT, Kozeowski TT (eds) Ecophysiology of tropical crops. Academic Press, New York, NY, pp. 113–156
Bartholomew DP, Malézieux EP (1994) Pineapple. In: Schaffer B, Andersen PC (eds) Handbook of environmental physiology of fruit crops, vol 2. CRC Press, Boca Raton, pp. 243–291
Bartholomew DP, Paull RE, Rohrbach KG (eds) (2002) The pineapple: botany, production, and uses. CABI, Wallingford
Barz M, Delivand MK (2011) Agricultural residues as promising biofuels for biomass power generation in Thailand. J Sustainable Energy Environment Special Issue 2011:21–27
Burg SP, Burg EA (1966) Auxin-induced ethylene formation: its relation to flowering in the pineapple. Science 152:1269–1269
Di Scala F, Dupuis L, Gaiddon C, De Tapia M, Jokic N, Gonzalez de Aguilar JL, Raul JS, Ludes B, Loeffler JP (2005) Tissue specificity and regulation of the N-terminal diversity of reticulon 3. Biochem J 385(Pt 1):125–134
Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8:967–974
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35:W297–W299
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Lind MI, Ekengren S, Melefors Ö, Söderhäll K (1998) Drosophila ferritin mRNA: alternative RNA splicing regulates the presence of the iron-responsive element. FEBS Lett 436:476–482
Lum G, Meinken J, Orr J, Frazier S, Min XJ (2014) PlantSecKB: the plant secretome and subcellular proteome knowledgebase. Comput Molec Biol 4:1–17
Li J, Li X, Guo L, et al. (2006) A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J Exp Bot 57:1263–1273
Lv L, Duan J, Xie J, Wei C, Liu Y, Liu S, Sun G (2012a) Isolation and characterization of a FLOWERING LOCUS T homolog from pineapple (Ananas comosus (L.) Merr). Gene 505:368–373
Lv LL, Duan J, Xie JH, Liu YG, Wei CB, Liu SH, Zhang JX, Sun GM (2012b) Cloning and expression analysis of a PISTILLATA homologous gene from pineapple (Ananas comosus L. Merr). Int J Mol Sci 13:1039–1053
Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22:1184–1195
McCarthy FM, Wang N, Magee GB, Williams WP, Luthe DS, Burgess SC (2006) AgBase: a functional genomics resource for agriculture. BMC Genomics 7:229
Min X, Bartholomew DP (1996) Effect of plant growth regulators on ethylene production, 1-aminocyclopropane-1-carboxylic acid oxidase activity, and initiation of inflorescence development of pineapple. J Plant Growth Regul 15:121–128
Min XJ, Powell B, Braessler J, Meinken J, Yu F, Sablok G (2015) Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops. BMC Genomics 16:721
Min XJ, Butler G, Storms R, Tsang A (2005a) OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res 33:W677–W680
Min XJ, Butler G, Storms R, Tsang A (2005b) TargetIdentifier: a web server for identifying full-length cDNAs from EST sequences. Nucleic Acids Res 33:W669–W672
Min XJ (2013) ASFinder: a tool for genome-wide identification of alternatively spliced transcripts from EST-derived sequences. Int J Bioinforma Res Appl 9:221–226
Ming R, VanBuren R, Wai CM, et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet. doi:10.1038/ng.3435
Morello L, Breviario D (2008) Plant spliceosomal introns: not only cut and paste. Curr Genet 9:227–238
Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR (2005) Developing pineapple fruit has a small transcriptome dominated by metallothionein. J Exp Bot 56:101–112
Nievola CC, Kraus JE, Freschi L, Souza BM, Mercier H (2005) Temperature determines the occurrence of CAM or C3 photosynthesis in pineapple plantlets grown in vitro. In Vitro Cellular Dev Biol-Plant 41:832–837
Nziengui H, Bouhidel K, Pillon D, Der C, Marty F, Schoefs B (2007) Reticulon-like proteins in Arabidopsis Thaliana: structural organization and ER localization. FEBS Lett 581:3356–3362
Ong WD, Voo LYC, Kumar VS (2012) De novo assembly, characterization and functional annotation of pineapple fruit transcriptome through massively parallel sequencing. PLoS One 7:e46937
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415
Reddy AS, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25:3657–3683
Sablok G, Gupta PK, Baek JM, Vazquez F, Min XJ (2011) Genome-wide survey of alternative splicing in the grass Brachypodium Distachyon: an emerging model biosystem for plant functional genomics. Biotechnol Lett 33:629–636
Sablok G, Harikrishna JA, Min XJ (2013) Next generation sequencing for better understanding alternative splicing: way ahead for model and non-model plants. Transcriptomics 1:e103
Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T (2014) Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci U S A 111:18781–18786
Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25:3640–3656
Surles T, Foley M, Turn S, Staackmann M (2009) A scenario for accelerated use of renewable resources for transportation fuels in Hawai ‘i. University of Hawaii, Hawaii Natural Energy Institute, School of Ocean and Earth Science and Technology, pp. 1–38
Taussig SJ, Batkin S (1988) Bromelain, the enzyme complex of pineapple (Ananas comosus) and its clinical application: an update. J Ethnopharmacol 22:191–203
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc 7:562–578
Trusov Y, Botella JR (2006) Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.]. J Exp Bot 57:3953–3960
VanBuren R, Walters B, Ming R, Min XJ (2013) Analysis of expressed sequence tags and alternative splicing genes in sacred lotus (Nelumbo Nucifera Gaertn.). Plant Omics J 6:311–317
Walters B, Lum G, Sablok G, Min XJ (2013) Genome-wide landscape of alternative splicing events in Brachypodium Distachyon. DNA Res 20:163–171
Wang B, Brendel V (2006) Genome wide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci U S A 103:7175–7180
Wang RH, Hsu YM, Bartholomew DP, Maruthasalam S, Lin CH (2007) Delaying natural flowering in pineapple through foliar application of aviglycine, an inhibitor of ethylene biosynthesis. HortSci 42:1188–1191
Yang YS, Strittmatter SM (2007) The reticulons: a family of proteins with diverse functions. Genome Biol 8:234
Zancani M, Peresson C, Biroccio A, Federici G, Urbani A, Murgia I, et al. (2004) Evidence for the presence of ferritin in plant mitochondria. Eur J Biochem 271:3657–3664
Zhang J, Liu J, Ming R (2014) Genomic analyses of the CAM plant pineapple. J Exp Bot 65:3395–3404
Zhao C, Beers E (2013) Alternative splicing of Myb-related genes MYR1 and MYR2 may modulate activities through changes in dimerization, localization, or protein folding. Plant Signal Behav 11:e27325
Acknowledgments
The work was supported by the University of Illinois at Urbana-Champaign to RM and Youngstown State University to XJM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Paulo Arruda
Electronic supplementary material
Supplementary Table 1
Alternative splicing events in pineapple (XLSX 578 kb)
Supplementary Table 2
Protein family (Pfam) distribution of proteins encoded by genes undergoing alternative splicing and genes not undergoing alternative splicing in pineapple (XLSX 193 kb)
Supplementary Table 3
Protein domain changes in different isoforms of alternatively spliced genes (XLSX 77 kb)
Supplementary Table 4
Alternative splicing isoforms encoded proteins having different subcellular locations (XLSX 20 kb)
Supplementary Table 5
Differentially expressed genes in pineapple (XLSX 142 kb)
Supplementary Table 6
Lists of differentially expressed genes (DEG), genes having transcripts alternatively spliced (AS), and genes with both DEG and AS (XLSX 119 kb)
Rights and permissions
About this article
Cite this article
Wai, C.M., Powell, B., Ming, R. et al. Analysis of Alternative Splicing Landscape in Pineapple (Ananas comosus). Tropical Plant Biol. 9, 150–160 (2016). https://doi.org/10.1007/s12042-016-9168-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-016-9168-1