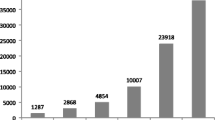
Abstract
It has become clear that dispersed repeat sequences have played multiple roles in eukaryotic genome evolution including increasing genetic diversity through mutation, inducing changes in gene expression, and facilitating generation of novel genes. Growing recognition of the importance of dispersed repeats has fueled development of computational tools designed to expedite discovery and classification of repeats. Here we review major existing repeat exploration tools and discuss the algorithms utilized by these tools. Special attention is devoted to ab initio programs, i.e., those tools that do not rely upon previously identified repeats to find new repeat elements. We conclude by discussing the strengths and weaknesses of current tools and highlighting additional approaches that may advance repeat discovery/characterization.

Similar content being viewed by others
Notes
Haas and Salzberg [33] have recently reviewed a subset of the repeat finders that we discuss. The focus of much of their review is mechanisms for handling the complications presented by repeats during genome assembly. The focus of our review is the use of these tools for identification of novel dispersed repeats in genomes.
BLAST is an acronym for the “Basic Local Alignment and Search Tool” developed by Altschul et al. [3]. There are currently several different BLAST modules specially designed for comparisons between different data types (see http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). WU-BLAST is a powerful alternative implementation of BLAST available from Washington University (http://blast.wustl.edu/). Crossmatch is a similarity search tool traditionally packaged with Phrap (www.phrap.org).
Abbreviations
- BLAST:
-
Basic Local Alignment and Search Tool
- bp:
-
base pair
- Mb:
-
megabase
- Gb:
-
gigabase
- MITE:
-
miniature inverted-repeat transposable element
- PALS:
-
Pairwise Alignment of Long Sequences
- SSR:
-
simple sequence repeat
References
Abouelhoda MI, Kurtz S, Ohlebusch E (2004) Replacing suffix trees with enhanced suffix arrays. J Discrete Algorithm 2:53–86
Agarwal P, States DJ (1994) The Repeat Pattern Toolkit (RPT): analyzing the structure and evolution of the C. elegans genome. Proc Int Conf Intell Syst Mol Biol 2:1–9
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Zhang J et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andrieu O, Fiston AS, Anxolabehere D et al (2004) Detection of transposable elements by their compositional bias. BMC Bioinformatics 5:94
Assaad FF, Tucker KL, Signer ER (1993) Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol 22:1067–1085
Bao Z, Eddy SR (2002) Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12:1269–1276
Batzer MA, Deininger PL (2002) ALU repeats and human genomic diversity. Nature 3:370–380
Bennett MD, Leitch IJ (2004) Plant DNA C-values database (release 3.0, Jan. 2004). http://www.rbgkew.org.uk/cval/homepage.html
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Biemont C, Vieira C (2006) Genetics: junk DNA as an evolutionary force. Nature 443:521–524
Britten RJ (1996) Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol Phylogenet Evol 5:13–17
Britten RJ, Kohne DE (1968) Repeated sequences in DNA. Science 161:529–540
Brosius J (2003) How significant is 98.5% ‘junk’ in mammalian genomes. Bioinformatics 19(suppl. 2):ii35
Campagna D, Romualdi C, Vitulo N et al (2005) RAP: a new computer program for de novo identification of repeated sequences in whole genomes. Bioinformatics 21:582–588
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Chenna R, Sugawara H, Koike T et al (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Chouvarine P, Saha S, Peterson DG (2008) An automated, high-throughput sequence read classification pipeline for preliminary genome characterization. Anal Biochem 373:78–87
Cormen TH, Leiserson CE, Rivest RL et al (2001) Introduction to Algorithms, 2nd Edition. MIT Press and McGraw-Hill, Cambridge, MA
Coward E, Drablos F (1998) Detecting periodic patterns in biological sequences. Bioinformatics 14:498–507
de Bruijn NG (1946) A combinatorial problem. Proc Koninklijke Nederlandse Akademie v Wetenschappen 49:758–764
Delcher AL, Kasif S, Fleischmann RD et al (1999) Alignment of whole genomes. Nucleic Acids Res 27:2369–2376
Delcher AL, Phillippy A, Carlton J et al (2002) Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30:2478–2483
Dorer DR, Henikoff S (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77:993–1002
Du L, Zhou H, Yan H (2007) OMWSA: detection of DNA repeats using moving window spectral analysis. Bioinformatics 23:631–633
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Edgar RC (2007) PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics 8:18
Edgar RC, Myers EW (2005) PILER: identification and classification of genomic repeats. Bioinformatics 21(Suppl 1):i152–i158
Feschotte C, Wessler SR (2001) Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes. Proc Natl Acad Sci USA 98:8923–8924
Frost LS, Leplae R, Summers AO et al (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732
Gusfield D (1999) Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press, New York
Haas BJ, Salzberg SL (2007) Finding repeats in genome sequences. In: Lengauer T (ed) Bioinformatics—From Genomes to Therapies, 1 edn. Wiley-VCH, Weinheim, pp 197–234
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225
Hou M, Berman P, Hsu CH et al (2007) HomologMiner: looking for homologous genomic groups in whole genomes. Bioinformatics 23:917–925
Ilie L, Ilie S (2007) Multiple spaced seeds for homology search. Bioinformatics 23:2969–2977
Jiang N, Bao Z, Zhang X et al (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431:569–573
Jiang N, Bao Z, Zhang X et al (2003) An active DNA transposon family in rice. Nature 421:163–167
Jurka J, Kapitonov VV, Pavlicek A et al (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Jurka J, Klonowski P, Dagman V et al (1996) CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20:119–121
Kalendar R, Vicient CM, Peleg O et al (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kapitonov VV, Jurka J (2001) Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A 98:8714–8719
Kapitonov VV, Jurka J (2006) Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A 103:4540–4545
Kolpakov R, Bana G, Kucherov G (2003) mreps: Efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res 31:3672–3678
Kurtz S, Choudhuri JV, Ohlebusch E et al (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642
Kurtz S, Schleiermacher C (1999) REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15:426–427
Lai J, Li Y, Messing J et al (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102:9068–9073
Lapitan NLV (1992) Organization and evolution of higher plant nuclear genomes. Genome 35:171–181
Lee C, Ritchie DBC, Lin CC (1994) A tandemly repetitive, centromeric DNA sequence from the Canadian woodland caribou (Rangifer tarandus caribou): its conservation and evolution in several deer species. Chromosome Res 2:293–306
Lefebvre A, Lecroq T, Dauchel H et al (2003) FORRepeats: detects repeats on entire chromosomes and between genomes. Bioinformatics 19:319–326
Li M, Ma B, Kisman D et al (2004a) Patternhunter II: highly sensitive and fast homology search. J Bioinform Comput Biol 2:417–439
Li R, Ye J, Li S et al (2005) ReAS: Recovery of ancestral sequences for transposable elements from the unassembled reads of a whole genome shotgun. PLoS Comput Biol 1:e43
Li X, Rao S, Wang Y et al (2004b) Gene mining: a novel and powerful ensemble decision approach to hunting for disease genes using microarray expression profiling. Nucleic Acids Res 32:2685–2694
Li YC, Korol AB, Fahima T et al (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465
Lundblad V, Wright WE (1996) Telomeres and telomerase: A simple picture becomes complex. Cell 87:369–375
Ma B, Tromp J, Li M (2002) PatternHunter: faster and more sensitive homology search. Bioinformatics 18:440–445
Mak D, Gelfand Y, Benson G (2006) Indel seeds for homology search. Bioinformatics 22:e341–e349
Manber U, Myers G (1993) Suffix arrays: a new method for on-line string searches. SIAM J Comput 22:935–948
McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Morgante M, Brunner S, Pea G et al (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37:997–1002
Müller HJ (1930) Types of viable variations induced by X-rays in Drosophila. Genetics 22:299–337
Nagl W (1976) DNA endoreduplication and polyteny understood as evolutionary strategies. Nature 261:614–615
Ohshima K, Okada N (2005) SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res 110:475–490
Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32:D360–D363
Pevzner PA, Tang H, Tesler G (2004) De novo repeat classification and fragment assembly. Genome Res 14:1786–1796
Price AL, Jones NC, Pevzner PA (2005) De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl 1):i351–i358
Pritham EJ, Putliwala T, Feschotte C (2007) Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390:3–17
Quesneville H, Bergman CM, Andrieu O et al (2005) Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol 1:166–175
Ruitberg CM, Reeder DJ, Butler JM (2001) STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acids Res 29:320–322
Saha S, Bridges S, Magbanua ZV et al. (2008) Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res (in press)
Sharma D, Issac B, Raghava GP et al (2004) Spectral Repeat Finder (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics 20:1405–1412
Sherman JD, Stack SM (1995) Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141:683–708
Smit AFA, Hubley R, Green P (1996–2004) RepeatMasker Open-3.0. http://www.repeatmasker.org
Sonnhammer ELL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:1–10
Sperber GO, Airola T, Jern P et al (2007) Automated recognition of retroviral sequences in genomic data—RetroTector©. Nucleic Acids Res 35:4964–4976
Strachan T, Read AP (1999) Human molecular genetics, 2nd edn. Wiley & Sons, New York
Syvanen M (1984) The evolutionary implications of mobile genetic elements. Annual Rev Genet 18:271–293
Tan AC, Gilbert D (2003) Ensemble machine learning on gene expression data for cancer classification. Appl Bioinformatics 2:S75–S83
Taneda A (2004) Adplot: detection and visualization of repetitive patterns in complete genomes. Bioinformatics 20:701–708
Temnykh S, DeClerck G, Lukashova A et al (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Timberlake WE (1978) Low repetitive DNA content in Aspergillus nidulans. Science 202:973–975
Toth G, Deak G, Barta E et al (2006) PLOTREP: a web tool for defragmentation and visual analysis of dispersed genomic repeats. Nucleic Acids Res 34:W708–W713
Tu Z (2001) Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A 98:1699–1704
Volfovsky N, Haas BJ, Salzberg SL (2001) A clustering method for repeat analysis in DNA sequences. Genome Biol 2:research0027.1–0027.11
Wang J, Wong GK, Ni P et al (2002) RePS: a sequence assembler that masks exact repeats identified from the shotgun data. Genome Res 12:824–831
Warburton PE, Giordano J, Cheung F et al (2004) Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res 14:1861–1869
Weiner P (1973) Linear pattern matching algorithm. In: Proceedings of the 14th annual IEEE symposium on switching and automata theory, University of Iowa, Iowa City, 15–17 Oct 1973
Wessler SR (1997) Transposable elements and the evolution of gene expression. Exp Biol 1039:115–122
Wicker T, Matthews DE, Keller B (2002) TREP: a database for Triticeae repetitive elements. Trends Plant Sci 7:561–562
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Yang G, Hall TC (2003) MAK, a computational tool kit for automated MITE analysis. Nucleic Acids Res 31:3659–3665
Zuckerkandl E, Hennig W (1995) Tracking heterochromatin. Chromosoma 104:75–83
Acknowledgements
This research was supported, in part, by the National Science Foundation (DBI-0421717 to D.G.P. and EPS-0556308 to S.M.B.), the United States Department of Agriculture (CSREES-2006-34506-17290 and ARS-58-6402-7-241 to D.G.P.), and the Mississippi Corn Promotion Board (to D.G.P.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Dr. Ray Ming
Rights and permissions
About this article
Cite this article
Saha, S., Bridges, S., Magbanua, Z.V. et al. Computational Approaches and Tools Used in Identification of Dispersed Repetitive DNA Sequences. Tropical Plant Biol. 1, 85–96 (2008). https://doi.org/10.1007/s12042-007-9007-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-007-9007-5