Abstract
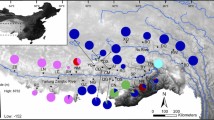
Trade and collection of edible frogs are banned in India. We used mitochondrial (16 and 12S DNA) and nuclear gene (Rag-1 and Rhodopsin) sequences to examine the population genetic and demographic structure of an edible frog species, Phrynoderma karaavali (Karaavali Skittering frog) from Kerala as it exist after the ban. Frogs from 11 sites show high mtDNA haplotype and nDNA diversity which indicates a stable or expanding population. The evolutionary demographic pattern suggests population expansion across its geographical range, even though the species is still subject to poaching. Two major population clusters were observed at the northern and southern end of the species range. Gene flow occurs despite of geographic barriers. Genetic distance increases with geographical distance. P. karaavali diverged from its sister species in Phrynoderma around 11 mya in the late Miocene.








Similar content being viewed by others
References
Abdulali H. 1985 On the export of frog legs from India. J. Bombay Nat. Hist. Soc. 82, 347–375.
Altherr S., Goyenechea A. and Schubert D. 2011 Canapés to extinction: the international trade in frogs’ legs and its ecological impact.–Pro Wildlife, Defenders of Wildlife. Animal Welfare Institute. Munich, Germany.
Anoop V., Kumar K. S., Sivakumar K., Reghunathan D., Manoj P., Deuti K. et al. 2017 The complete mitochondrial genome of Euphlyctis karaavali (Amphibia: Anura) with a note on its range expansion. Conserv. Genet. Resour. 9, 427–430.
Bandelt H. J., Forster P. and Röhl A. 1999 Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48.
Biju S. and Bossuyt F. 2003 New frog family from India reveals an ancient biogeographical link with the Seychelles. Nature 425, 711–714.
Bossuyt F. and Milinkovitch M. C. 2000 Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc. Natl. Acad. Sci. USA 97, 6585–6590.
Bossuyt F., Brown R. M., Hillis D. M., Cannatella D. C. and Milinkovitch M. C. 2006 Phylogeny and biogeography of a cosmopolitan frog radiation: Late Cretaceous diversification resulted in continent-scale endemism in the family Ranidae. Syst. Biol. 55, 579–594.
Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C. H., Xie D. et al. 2014 BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS. Comput. Biol. 10, 1–6.
Carr S. M. and Marshall H. D. 2008 Intraspecific phylogeographic genomics from multiple complete mtDNA genomes in Atlantic Cod (Gadus morhua): Origins of the ‘“Codmother”,’ Transatlantic Vicariance and Midglacial Population Expansion. Genetics 180, 381–389.
Chen G. Y., Wang B., Liu J. Y., Jiang J. P. and Gao P. 2019 Population genetic diversity of an odorous frog Odorrana grahami (Amphibia: Anura: Ranidae) in relation to conservation based on mitochondrial DNA. Mitochondrial DNA Part B 4, 57–61.
Cicek K., Ayaz D., Afsar M., Bayraksi Y., Peksen C. A., Cumhariyet O. et al. 2021 Unsustainable harvest of eater frogs in southern Turkey for the European market. Oryx 55, 364–372.
Daszak P., Berger L., Cunningham A. A., Hyatt A. D., Green D. E. and Speare R. 1999 Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5, 735–748.
de Castro Godinho M. B. and Da Silva F. R. 2018 The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Sci. Rep. 8, 1–11.
Dinesh K., Channakeshavamurthy B., Deepak P., Ghosh A. and Deuti K. 2021 Morphological groupings within Euphlyctis (Anura: Dicroglossidae) and description of a new species from the surroundings of Thattekad Bird Sanctuary, Kerala, India. Zootaxa 4990, 329–353.
Drummond A. J., Rambaut A., Shapiro B. and Pybus O. G. 2005 Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192.
Evanno G., Regnaut S. and Goudet J. 2005 Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620.
Excoffier L. and Lischer H. E. 2010 Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567.
Excoffier L., Smouse P. E. and Quattro J. M. 1992 Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Feng Y. J., Blackburn D. C., Liang D., Hillis D. M., Wake D. B., Cannatella D. C. et al. 2017 Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. 114, 5864–5870.
Fu Y. X. 1997 Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
Fugler C. 1983 The status of population of Rana tigrina Daudin in Bangladesh. Fish. Inform. Bull. Bangladesh 1, 1–5.
Gascon C. 2007 Amphibian conservation action plan: proceedings IUCN/SSC Amphibian Conservation Summit 2005 IUCN.
Gehring P. S., Pabijan M., Randrianirina J. E., Glaw F. and Vences M. 2012 The influence of riverine barriers on phylogeographic patterns of Malagasy reed frogs (Heterixalus). Mol. Phylogen. Evol. 64, 618–632.
George S. 1995 Status of Rana hexadactyla Lesson in Kerala. Zoos Print 10, 11–12.
George S. and Andrews M. I. 1995 Food and feeding habits of R. hexadactyla Lesson. J. Bombay Nat. Hist. Soc. 92, 220–224.
Gilbert M., Bickford D., Clark L., Johnson A., Joyner P. H., Keatts L. O. et al. 2012 Amphibian pathogens in Southeast Asian frog trade. EcoHealth 9, 386–398.
Goode M. J., Swann D. E. and Schwalbe C. R. 2004 Effects of destructive collecting practices on reptiles: a field experiment. J. Wildl. Manage 68, 429–434.
Govindaraju D. R. 1989 Variation in gene flow levels among predominantly self-pollinated plants. J. Evol. Biol. 2, 173–181.
Guillot G., Mortier F. and Estoup A. 2005 GENELAND: a computer package for land scape genetics. Mol. Ecol. Notes 5, 712–715.
Harikumar P. S. P., Deepak R. and Sabitha A. R. 2014 Water quality assessment of Valapattanam river basin in Kerala, India, using macro-invertebrates as biological indicators. Open Environ. Biol. Monit. J. 6, 1–9.
Heled J. and Drummond A. J. 2008 Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8, 1–15.
Heyer R., Donnelly M. A., Foster M. and Mcdiarmid R. 2014 Measuring and monitoring biological diversity: standard methods for amphibians. Smithsonian Institution Press, Washington.
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. et al. 2012 Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
Kimura M. 1980 A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Kumar K. S., Chandrika S. K. and George S. 2020 Genetic structure and demographic history of Indirana semipalmata, an endemic frog species of the Western Ghats, India. Mitochondrial DNA Part A 31, 365–378.
Kumar S., Stecher G., Li M., Knyaz C. and Tamura K. 2018 MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Kusrini M. D. 2005 Edible frog harvesting in Indonesia: evaluating its impacts and ecological context. (PhD dissertation). Queensland, Australia: James Cook University.
Li R., Chen W., Tu L. and Fu J. 2009 Rivers as barriers for high elevation amphibians: a phylogeographic analysis of the alpine stream frog of the Hengduan Mountains. J. Zool. 277, 309–316.
Librado P. and Rozas J. 2009 DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
Low V. L., Adler P. H., Takaoka H., Ya’cob Z., Lim P. E., Tan T. K. et al. 2014 Mitochondrial DNA markers reveal high genetic diversity but low genetic differentiation in the black fly Simulium tani Takaoka & Davies along an elevational gradient in Malaysia. PLoS One 9, 1–10.
Manni F., Guérard E. and Heyer E. 2004 Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum. Biol. 76, 173–190.
Mohneke M. 2011 (Un)sustainable use of frogs in West Africa and resulting consequences for the ecosystem. MSc thesis, Mathematisch-Naturwissenschaftliche Fakultät, Humboldt- Universität zu Berlin, Berlin, Germany.
Moraes L. J., Pavan D., Barros M. C. and Ribas C. C. 2016 The combined influence of riverine barriers and flooding gradients on biogeographical patterns for amphibians and squamates in south-eastern Amazonia. J. Biogeogr. 43, 2113–2124.
Nei M. 1983 Evolutionary change of amino acid sequences. In Molecular evolutionary genetics, pp 39-63. Columbia University Press, New York.
Niekisch M. 1986 The international trade in frogs’ legs. Traffic Bull. 8, 7–10.
Oza G. 1990 Ecological effects of the frog’s legs trade. Environmentalist 10, 39–42.
Palumbi S. R., Martin A. P., Romano S. L., McMillan W. O., Stice L. and Grabowski G. 1991 The simple fool’s guide to PCR. Department of Zoology, University of Hawaii, Honolulu.
Pandian T. and Marian M. P. 1986 Production and utilization of frogs: an ecological view. Proc. Anim. Sci. 95, 289–301.
Pidancier N., Miquel C. and Miaud C. 2003 Buccal swabs as a non-destructive tissue sampling method for DNA analysis in amphibians. Herpetol. J. 13, 175–178.
Pritchard J. K., Wen W. and Falush D. 2009 Documentation for STRUCTURE software: Version 2. University of Chicago, Chicago, IL, pp. 1-36.
Priti H., Naik C. R., Seshadri K. S., Singal R., Vidisha M. K., Ravikanth G., V. et al. 2016 A new species of Euphlyctis (Amphibia, Anura, Dicroglossidae) from the west coastal plains of India. Asian Herpetol. Res. 7, 229–241.
Raghavendra K., Sharma P. and Dash A. P. 2008 Biological control of mosquito populations through frogs: opportunities and constrains. Indian J. Med. Res. 128, 22–25.
Rambaut A., Drummond A. J., Xie D., Baele G. and Suchard M. A. 2018 Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901.
Ramos-Onsins S. E. and Rozas J. 2002 Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100.
Ranjeet K., Radhakrishnan K., Sureshkumar S. and Kurup B. M. 2012 Athropogenic threats to fish biodiversity of Periyar lake, A Western Ghats hotspot. Biodiversity: Utilization. Threats and Cultural Linkages 9, 65–79.
Reed D. H. and Frankham R. 2003 Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237.
Reed D. H. and Hobbs G. R. 2004 The relationship between population size and temporal variability in population size. Anim. Conserv. 7, 1–8.
Roelants K., Jiang J. and Bossuyt F. 2004 Endemic ranid (Amphibia: Anura) genera in southern mountain ranges of the Indian subcontinent represent ancient frog lineages: evidence from molecular data. Mol. Phylogen. Evol. 31, 730–740.
Rogers A. R. and Harpending H. 1992 Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569.
Schlaepfer M. A., Hoover C. and Dodd C. K. 2005 Challenges in evaluating the impact of the trade in amphibians and reptiles on wild populations. BioScience 55, 256–264.
Shields G. and Gust J. 1995 Lack of geographic structure in mitochondrial DNA sequences of Bering Sea walleye pollock, Theragra chalcogramma. Mol. Mar. Biol. Biotechnol. 4, 69–82.
Slatkin M. and Hudson R. R. 1991 Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562.
Smouse P. E., Long J. C. and Sokal R. R. 1986 Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35, 627–632.
Song J., Hou F., Zhang X., Yue B. and Song Z. 2014 Mitochondrial genetic diversity and population structure of a vulnerable freshwater fish, rock carp (Procypris rabaudi) in upper Yangtze River drainage. Biochem. Syst. Ecol. 55, 1–9.
Tajima F. 1989 Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tamura K., Battistuzzi F. U., Billing-Ross P., Murillo O., Filipski A. and Kumar S. 2012 Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 109, 19333–19338.
Wright S. 1949 The genetical structure of populations. Ann. Eugen. 15, 323–354.
Zhang Y. H., Zhao Y. Y., Li X. Y. and Li X. C. 2016 Evolutionary history and population genetic structure of the endemic tree frog Hyla tsinlingensis (Amphibia: Anura: Hylidae) inferred from mitochondrial gene analysis. Mitochondrial DNA Part A 27, 1348–1357.
Acknowledgements
We thank Dr Manoj P. (RGCB) for gene sequencing. We are grateful to Kerala Forest Department for permissions for sample collections (KFDHQ-28751/2018-CWW/WL10). We also thank RGCB for funding the work from the central grand. Anoop V. S. thank University Grants commission (UGC) for the research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Steven M. Carr
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anoop, V.S., George, S. Population genetic structure and evolutionary demographic patterns of Phrynoderma karaavali, an edible frog species of Kerala, India. J Genet 102, 8 (2023). https://doi.org/10.1007/s12041-022-01407-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-022-01407-5