Abstract
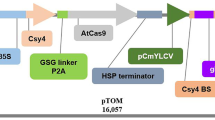
CmFT homologous gene in muskmelon was obtained by homologous cloning, introducing CmFT gene by Agrobacterium-mediated transformation. The results of subcellular localization showed that CmFT protein was expressed in cytoplasm and nucleus. qRT-PCR results showed that the expression levels of AtLFY, AtFT, AtCO, AtFLC, AtSOC1 and AtAP1 were upregulated in the 35S::MeFT Arabidopsis line. The CmFT gene was introduced into wild-type Arabidopsis by Agrobacterium-mediated transformation, and the growth status of T2 transgenic Arabidopsis thaliana and wild-type A. thaliana was observed. The results showed that wild-type Arabidopsis began to bolt on the 25th day after sowing, we can initially confirm that the FT gene of melon can promote the early flowering of melon in the growth and development of melon.







Similar content being viewed by others
References
Adeyemo O. S., Chavarriaga P., Tohme J., Fregene M., Davis S. J. and Setter T. L. 2017 Overexpression of Arabidopsis FLOWERING LOCUS T (FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PLoS One 12.
Balasubramanian S. and Weigel D. 2006 Temperature induced flowering in Arabidopsis thaliana. Plant Signal. Behav. 1, 227–228.
Coelho C. P., Mark A. M., Antonio C. J. and Joseph C. 2014 Putative sugarcane FT/TFL1 genes delay flowering time and alter reproductive architecture in Arabidopsis. Front. Plant Sci. 5, 221.
Fan C. M., Hu R. B., Zhang X. M., Wang X., Zhang W. J., Zhang Q. Z. et al. 2014 Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 14, 9.
Hsu C. Y., Adams J. P., Kim H. J., No K., Ma C. P., Strauss S. H. et al. 2011 FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 108, 10756–10761.
Imamura T., Nakatsuka T., Higuch A., Nishihara M. and Takahashi H. 2011 The gentian orthologs of the FT/TFL1 gene family control floral initiation in Gentiana. Plant Cell Physiol. 52, 1031–1041.
Jiang D., Liang J. L., Chen X. L., Hong B., Jia W. S. and Zhao L. J. 2010 Transformation of Arabidopsis flowering gene FT to from Cut Chrysanthemum ‘Jinba’ by Agrobacterium Mediate. Hortic. Sin. 37, 441–448.
Kikuchi R., Kawahigashi H., Ando T., Tonooka T. and Handa H. 2009 Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353.
Kong F. J., Liu B. H., Xia Z. J., Sato S., Kim B. M., Watanabe S. et al. 2010 Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231.
Kotoda N., Hayashi H., Suzuki M., Igarash M., Hatsuyama Y., Kidou S. et al. 2010 Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malusdomestica Borkh.). Plant. Cell. Physiol. 51, 561–575.
Książkiewicz M., Rychel S., Nelson M. N., Wyrwa K., Naganowska B. and Wolko B. 2016 Expansion of the phosphatidylethanolamine binding protein family in legumes: a case study of Lupinus angustifolius L. FLOWERING LOCUS T homologs, LanFTc1 and LanFTc2. BMC Genome 17, 820.
Lee R., Baldwin S., Kenel F., McCallum J. and Macknight R. 2013 FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 4, 2884.
Liu K. D., Feng S. X., Pan Y. L., Li H. L., Chen Y. and Yuan C. C. 2016 Cloning and expression analysis of CpFT1 Gene and its promoter from Papaya. Hortic. Sin. 43, 2359–2368.
Li X. F., Jia L. Y., Xu J., Deng X. J., Wang Y., Zhang W. et al. 2013 FT-like NFT1 gene may play a role in flower transition induced by heat accumulation in Narcissus tazetta var. chinensis. Plant Cell Physiol. 54, 270–281.
Li W. M., Wang S. S., Yao Y. X., Zhao C. Z., Hao Y. J. and You C. X. 2009 Genetic Transformation of Apple MdFT gene into Tomato. Hortic. Sin. 36, 1255–1260.
Lv Y. J., Wang C. X., Yang W. J. and Zhang Y. S. 2015 FT homologous gene in higher plants: progress on genes and mechanisms. Mol. Plant Breed. 13, 1415–1423.
Nakano Y., Higuchi Y., Sumitomo K. and Hisamatsu T. 2013 Flowering retardation by high temperature in chrysanthemums: involvement of FLOWERING LOCUS T-like 3 gene repression. J. Exp. Bot. 64, 909–920.
Nan H. Y., Cao D., Zhang D. Y., Li Y., Lu S. J., Tang L. L. et al. 2014 GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One 9, e97669.
Rosas U., Mei Y., Xie Q., Banta J. A., Zhou R. W., Eufferheld G. et al. 2014 Variation in Arabidopsis flowering time associated with cisregulatory variation in CONSTANS. Nat. Commun. 5, 3651.
Shen L. L., Chen Y., Su X. H., Zhang S. G., Pan H. X. and Huang M. 2012 Two FT orthologs from Populus simonii Carrière induce early flowering in Arabidopsis and poplar trees. Plant Cell Tiss. Org. Cult. 108, 371–379.
Shim J. S., Kubota A. and Imaizumi T. 2017 Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15.
Song G., Aaron W., Zhao D. Y., Jiang N. and James F. H. 2013 The Vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): a flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Rep. 32, 1759–1769.
Sun H. B., Jia Z., Cao S., Jiang B. J., Wu C. X., Hou W. S. et al. 2011 GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One 6, e29238.
Wu L., Liu D., Wu J., Zhang R., Qin Z., Liu D. et al. 2013 Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell. 25, 4363–4377.
Yasushi K. and Detlef W. 2007 Move on up, it’s time for change-mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384.
Yuan M., Xing C. B., Ge W. N., Wang L. and Guo D. 2017 Protein expression and purification of flowering inducer gene FT in Arabidopsis thaliana. Genom. Appl. Biol. 36, 3053–3056.
Zhang J. Yan S. S., Zhao W. S. and Zhang X. L. 2013 Cloning and functional analysis of CsFT in Cucumber. Hortic. Sin. 40, 2180–2188.
Zhang F., Xin M., Yu S. Q., Liu D., Zhou X. Y. and Qin Z. W. 2019 Expression and functional analysis of the propamocarb-related gene CsMCF in cucumber. Front. Plant Sci. 10, 871.
Zhou S. S., Jiang L., Guan S. X., Gao Y. X., Gao Q. H., Wang G. D. et al. 2018 Expression profiles of five FT-like genes and functional analysis of PhFT-1 in a Phalaenopsis hybrid. Electron. J. Biotechnol. 31, 75–83.
Zhou Q., Zhang S. S., Bao M. Z. and Liu G. F. 2018 Advances on molecular mechanism of floral initiation in higher plants. Mol. Plant Breed. 16, 3681–3692.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31640069), project from Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Northeast Region), and project from Ministry of Agriculture/Northeast Agricultural University (201603). Anhui Provincial University Super Talents Funding Programme (gxgwfx2018033), partly supported by the open funds of the State Key Laboratory of Crop Genetics and Germplasm Enhancement (ZW201708).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Rights and permissions
About this article
Cite this article
Zhang, H., Zhang, Y. Molecular cloning and functional characterization of CmFT (FLOWERING LOCUS T) from Cucumis melo L.. J Genet 99, 41 (2020). https://doi.org/10.1007/s12041-020-1191-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-020-1191-1